Abstract
This study aimed to understand the expression of solute carrier family 12 member 8 (SLC12A8) in breast carcinoma and its biological functions, as well as its effect on the Toll-like receptor /NOD-like receptor (TLR/NLR) signaling pathway. The expression of SLC12A8 was analyzed using the public RNA sequencing dataset from TCGA database and the two datasets from Oncomine database. The former dataset was also used to evaluate the prognostic value of SLC12A8 in breast carcinoma. Real-time qPCR and western blot were applied to measure relative expression of SLC12A8. Functionally, the effect of SLC12A8 on the cells proliferation and motion was studied using cell counting kit 8 and Transwell assays respectively. Mechanistic studies were conducted using Gene Set Enrichment Analysis (GSEA) and confirmed by western blot. As a result, SLC12A8 was upregulated in breast carcinoma, and high levels of SLC12A8 led to a poorer prognosis and can be regarded as an independent prognosticator for patients with breast carcinoma. Functional experiments demonstrated that SLC12A8-knockdown suppressed while SLC12A8-overexpression elevated the viability, invasiveness and motility of breast carcinoma cells. Furthermore, GSEA indicated that high SLC12A8 was positively correlated with TLR/NLR signaling pathway. Silencing SLC12A8 significantly reduced the protein expression of TLR/NLR-related markers, whereas overexpression of SLC12A8 caused an elevation on the protein expression of these markers. All these data suggested that SLC12A8 plays a promoting effect on the cells viability, invasiveness and motility in breast carcinoma by activating TLR/NLR signaling pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast carcinoma is the most frequently occurring cancer in women (Tao et al. 2015). Globally, an estimated 279,100 new cases of breast carcinoma will be diagnosed in 2020, accounting for 30% of all cancers among women (Siegel et al. 2020). Although metastasis and mortality from breast carcinoma have declined with advances in cancer treatment modalities, distant metastasis is still responsible for the deaths of breast carcinoma (Weigelt et al. 2005). Distant metastasis involved in a variety of complex mechanisms, including invasion and migration, angiogenesis, epithelial mesenchymal transformation etc. (Kozlowski et al. 2015). At present, the underlying mechanism of breast carcinoma pathogenesis and tumor metastasis has not been fully elucidated. Thus, full elucidation of the underlying mechanisms and identification of new effective therapeutic targets are of great significance to improve breast carcinoma survival.
The electroneutral cation-chloride cotransporter gene family, SLC12, was identified initially at the molecular level in fish and then in mammals, and contains nine members in vertebrates (Hebert et al. 2004). It plays a key role in the physiological processes such as cell volume regulation, transepithelial ion movement, intracellular chloride ion concentration regulation, and blood pressure regulation (Arroyo et al. 2013). Numerous physiological, immunohistochemical and biochemical studies have revealed the importance of this gene family to human health and disease (Gagnon and Delpire 2013). In cancers, it was also found that the members of SLC12 gene family are involved in the progression. A study showed that SLC12A5 is upexpressed in colorectal cancer, and could significantly promote xenograft tumor growth and lung metastasis (Xu et al. 2016). Through bioinformatics analysis from Gene Expression Omnibus database, SLC12A1 was found to be overexpressed in hepatocellular carcinoma Hep3B cells and treatment with the SLC12A1 antagonist could delay tumor formation and reduce Hep3B cell tumor size in mouse xenografts (Teng et al. 2016). Moreover, SLC12A2 was found to be upregulated in human colon cancer tissues using L linear ion trap Fourier transform mass spectrometry technology (Zhang et al. 2017). All these findings suggest that some members of SLC12 gene family may also be involved in the progression of breast carcinoma, which is rarely reported. Solute carrier family 12 member 8 (SLC12A8), also known as CCC9, belongs to the SLC12 gene family of the electroneutral cation-chloride-coupled cotransporters, and no physiological role has been ascribed to the protein encoded by this gene (Gagnon and Delpire 2013). Recently, the role of SLC12A8 in cancers has emerged, as reported that low mRNA expression of SLC12A8 were associated with decreased disease-free survival in pancreatic ductal adenocarcinoma (Feigin et al. 2017). In Seoul breast carcinoma study, the gene-based results suggested that SLC12A8 was associated with the overall survival, reminding that SLC12A8 might play a role in the progression in breast carcinoma (Kim et al. 2018). However, the knowledge of its functional roles and the underlying mechanisms in breast carcinoma are still limited.
Hence, the objective of this report was to evaluate the prognostic role of SLC12A8 in breast carcinoma based upon the downloadable datasets, and examine the influences of SLC12A8 on the phenotypes of breast carcinoma cells by loss- and gain-of-function of SLC12A8 assays, as well as explore the underlying mechanisms. All the data demonstrated that SLC12A8 shows diagnostic and prognostic values in breast carcinoma, and may have potential to be a considerable biomarker for the therapy of breast carcinoma.
Materials and methods
Microarray analysis
In current study, the breast infiltrating ductal carcinoma RNA-Seq data including 1109 breast carcinoma samples and 113 adjacent non-tumor samples were downloaded from The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov) database, and were used to analyze the expression pattern of SLC12A8 and assess its prognostic value. The correlation between SLC12A8 level and overall survival status was determined by Kaplan–Meier methods followed by a log-rank test. Cox univariate and multivariate analyses were used to identify independent predicators having a significant impact on the overall survival of patients with breast carcinoma. Gene Set Enrichment Analysis (GSEA) was conducted using GSEA 3.0 (http://www.broadinstitute.org/gsea/) to identify SLC12A8 related gene sets.
Cell culture and transfection
Human breast carcinoma cell lines MCF7 and Bcap-37, and normal cell line MCF-10A were purchased from Shanghai Cell Bank of Chinese Academy of Sciences. The cell lines were maintained in Roswell Park Memorial Institute medium 1640 (RPMI-1640) supplemented with 10% fetal calf serum (FBS) and antibiotics at 37 °C in 5% CO2.
SLC12A8 small interference RNAs (si-SLC12A8-1, 5′-GCAGGTGTCAAATGGATAA-3′; si-SLC12A8-2, 5′-CCATGTATATCACCGGCTT-3′) were transfected into MCF7 cells using Lipofectamine 2000 (Thermo Fisher Scientific, Carlsbad, USA) following manufacturer’s protocol to downregulate the expression level of SLC12A8. A scrambled siRNA (si-con, 5′-AATTCTCCGAACGTGTCACGT-3′) was used as their negative control. Plasmid pcDNA3.1-SLC12A8 was transfected into Bcap-37 cells to upregulate the expression level of SLC12A8 and using pcDNA3.1 empty vector as control. All the siRNAs and plasmids were synthesized by Shanghai GeneChem Corporation (Shanghai, China). 24 h after transfection, the cells were collected to measure the transfection efficiency and conduct the follow-up experiments.
RNA isolation and real-time qPCR analysis
Whole RNA was extracted from cells using the TRIzol Reagent (Takara, Kusatsu, Japan) according to the manufacturer’s instructions. The cDNA was reverse transcribed from 1 µg of total RNA by Reverse Transcription Kit (TaKaRa). Real-time qPCR was performed to evaluate the expression of SLC12A8 using a SYBR Premix Ex Taq (TaKaRa) in an Applied Biosystem 7500 fast detection system (Thermo Fisher Scientific, Carlsbad, USA). Quantitative data were normalized by GAPDH. The primers used in this study are listed below:
SLC12A8
F: 5′-TGGCGTCTACTCCATGATCTCC-3′,
R: 5′-CCGAGATGGATTCAGCAAAGCC-3′;
GAPDH
F: 5′-TAGATGACACCCGTCCCTGA-3′,
R: 5′-ACCTCCACCTGTCCTTAGTG-3′.
Western blot analysis
24 h after transfection, total protein was extracted from cells using a radioimmunoprecipitation (RIPA) assay buffer (Beyotime, Nantong, China) containing protease inhibitors. A bicinchoninic acid (BCA) assay (Thermo Fisher Scientific) served to quantify protein. Extracted proteins (20 µg per well) were added to the electrophoresis chambers and separated with 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto polyvinylidene fluoride (PVDF) membranes. Next, the membranes were blocked with 5% non-fat dried milk for an hour, then probed with the appropriate primary antibodies at 4 °C overnight. Then, these membranes were rinsed with Tris Buffered Saline Tween (TBST), and incubated with secondary antibodies for an hour at room temperature. After being washed in TBST thrice, the membranes were visualized using the enhanced chemiluminescence detection system (Thermo Fisher Scientific). And ImageJ software was applied to quantify the bands.
Antibodies used in this study including anti-SLC12A8 (PA5-107021), anti-TLR4 (48-2300), anti-MyD88 (PA5-19919), anti-phosphorylated (p)-NF-κB (PA5-17385), anti-NF-κB (PA5-82349), and anti-NOD2 (PA5-83584) were obtained from Thermo Fisher Scientific Corporation, while others antibodies including anti-NOD1 (#3545), anti-NLRP3 (#15,101), and anti-IL-1β (#12,703) were acquired from Cell Signaling Technology Corporation (Danvers, USA). Moreover, anti-p-IκBα (AF5821) and anti- IκBα (AF5204) were acquired from Beyotime Corporation (Nantong, China) and anti-pro-caspase 1 (ab179515) was acquired form Abcam (Cambridge, UK).
Cell proliferation assay
Breast carcinoma cells proliferation was evaluated using the cell counting kit 8 (CCK8, Beyotime, Nantong, China) following the manufacturer’s instructions. 24 h after transfection, 1 × 103 cells were seeded into a 96-well plate. After incubation for 24, 48, and 72 h, 10 µl CCK8 reagent was added to the culture medium, and incubation for another 1.5 h. The optical density (OD) values of each sample were recorded under a microplate reader at 450 nm.
Cell invasion and migration assay
Transwell invasion assay was implemented using 24-well Transwell chambers (Corning, NY, USA). 24 h after transfection, cells were suspended in 100 µl serum-free medium. Next, 1 × 104 cells were added to the upper chamber which was pre-coated with Matrigel. The lower chamber was filled with 500 µl medium plus with 10% FBS. Upon incubation for 24 h at 37 °C, cells that traversed the filter were rinsed with PBS, fix with 4% paraformaldehyde for 30 min and stained using crystal violet for 20 min. Finally, the cells were photographed under a fluorescence microscope in five random fields, and the number of cells was counted. The procedure of Transwell migration assay was similar to the invasion assay, except that the Transwell membranes were non-coated with Matrigel, and 0.5 × 104 cells were added to the upper chamber. These experiments were carried out for three independent times and data were expressed as mean ± standard deviation.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6.0 software (La Jolla, CA, USA) and SPSS 22.0 software (Armonk, NY, USA). The difference of double-group was analyzed using t test (two tailed), and one-way analysis of variance (ANOVA) was used to analyze the difference of multiple groups, followed by Tukey’s post hoc test. All experiments were performed at least three times, with one representative experiment shown, and the data were expressed as mean ± standard deviation. A value of p less than 0.05 was considered significant difference.
Results
SLC12A8 is upregulated in breast carcinoma
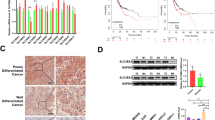
To help understand the role of SLC12A8 in breast carcinoma, we firstly analyzed its expression according to the RNA-Seq dataset from TCGA database, and found that the levels of SLC12A8 were significantly higher in breast carcinoma tissues than that in adjacent non-tumor tissues (p < 0.01, Fig. 1a). Similar results were obtained in the two datasets from Oncomine database when compared with normal breast tissues (p < 0.01, Fig. 1b, c). Next, to verify the transcriptional expression level of SLC12A8 in breast carcinoma, real-time qPCR analysis were performed in two commonly used cell lines (Bcap-37 and MCF7) of breast carcinoma. Results shown in Fig. 1d revealed that the expression of SLC12A8 was upregulated in two tumor cell lines compared with normal breast cell line MCF-10A (p < 0.01). All these data suggested that abnormal expression of SLC12A8 might play a role in the progression of breast carcinoma.
SLC12A8 was significantly upregulated in breast carcinoma and associated with unfavorable prognosis in patients with breast carcinoma. SLC12A8 expression in breast carcinoma tissues and relatively adjacent normal breast tissues in RNA-Seq dataset from TCGA database (a), in Curtis Breast dataset from Oncomine database (b) and in Richardson Breast 2 dataset from Oncomine database (c) was analyzed. d SLC12A8 expression in two breast carcinoma cell lines MCF7 and Bcap-37 was determined by real-time qPCR with normal breast cell line MCF-10A as control. e Kaplan-Meier analysis revealed that patients with low SLC12A8 expression (n = 506) had a better overall survival when compared with patients with high SLC12A8 expression group (n = 506). **p < 0.01
Expression of SLC12A8 shows an independent prognostic value
To further explore the prognostic value of SLC12A8 in breast carcinoma, a total of 1012 cases were divided into high and low SLC12A8 expression groups on the basis of the median expression of SLC12A8 using the RNA-Seq dataset from TCGA database. Kaplan-Meier curve showed that patients with low SLC12A8 expression had a higher probability of a better overall survival rate compared with high SLC12A8 expression (p < 0.05, Fig. 1e). In univariate analysis for breast carcinoma, SLC12A8 expression and clinical features including stage, tumor, node, metastasis, and age had prognostic values for overall survival (p < 0.05, Supplementary Table 1). Furthermore, Cox multivariate analysis demonstrated that SLC12A8 expression, stage, metastasis, and age were independent prognostic predictors for overall survival of breast carcinoma patients (p < 0.05, Supplementary Table 1). All these analyses suggested that SLC12A8 may serve as a considerable biomarker for the diagnosis and prognosis in breast carcinoma.
SLC12A8 enhances the proliferation of breast carcinoma cells
To generate better results for the functional experiments, MCF7 and Bcap-37 cells were chose to conduct knockdown and overexpression of SLC12A8 assays, respectively, due to their relatively high and low expression of SLC12A8 in the previous real-time qPCR analysis. The transfection efficiency of MCF7 cells treated with si-SLC12A8 and Bcap-37 cells treated with pcDNA3.1-SLC12A8 was measured by real-time qPCR and western blot. The outcomes showed that, compared with control or si-con group, the mRNA and protein expression of SLC12A8 were dramatically reduced when transfected with si-SLC12A8-1 or si-SLC12A8-2 (p < 0.01, Fig. 2a, b). Whereas SLC12A8 expression was significantly elevated after transfected with pcDNA3.1-SLC12A8 compared with control or vector group (p < 0.01, Fig. 2c, d). These detections suggested that we successfully established MCF7 cells with depleted SLC12A8 and Bcap-37 cells with overexpressed SLC12A8.
The effect of loss- and gain-of-function of SLC12A8 on the breast carcinoma cells proliferation. a, b Following treatment with con, si-con, si-SLC12A8-1, and si-SLC12A8-2 in MCF7 cells, the mRNA and protein expression levels of SLC12A8 were determined by real-time qPCR (a) and western blot (b) analyses. ##p < 0.01 vs. con group, **p < 0.01 vs. si-con group. c, d Following treatment with con, pcDNA3.1 vector, and pcDNA3.1-SLC12A8 in Bcap-37 cells, the mRNA and protein expression levels of SLC12A8 were determined by real-time qPCR (c) and western blot (d) analyses. ##p < 0.01 vs. con group, **p < 0.01 vs. si-con group. e, f Following treatment with si-con and si-SLC12A8-2 in MCF7 cells (e) or treatment with pcDNA3.1 vector and pcDNA3.1-SLC12A8 in Bcap-37 cells (f), the optical density values were determined by CCK8 assays. **p < 0.01
The viability (OD at 450 nm) of MCF7 and Bcap-37 cells was determined by CCK8 proliferation assay. After silencing SLC12A8, the MCF7 cells appeared a significant reduction of the OD values at 48 h and 72 h points in comparison with si-con group (p < 0.01, Fig. 2e, Supplementary Fig A), whereas Bcap-37 cells showed significantly higher OD values after overexpressing SLC12A8 compared with vector group (p < 0.01, Fig. 2f), suggesting that SLC12A8 may promote the cells proliferation in breast carcinoma.
SLC12A8 enhances the invasion and migration of breast carcinoma cells
For a deeper understanding of the functional role of SLC12A8 in breast carcinoma, Transwell invasion and migration assay were implemented. The results showed that SLC12A8-2-depleted MCF7 cells showed a dramatical reduction of the number in invasion and migration assays by contrast to their relative si-con group (p < 0.01, Fig. 3a, b, invasion: 126.67 ± 13.65 vs. 485.67 ± 24.01; migration: 216 ± 15.52 vs. 529.33 ± 24.42). Besides, MCF7 cells transfected with si-SLC12A8-1 showed the same trends, as observed that the invasive number in si-SLC12A8-1 group vs. si-con was 179.67 ± 28.5 vs. 299 ± 18.73, and the migratory number in si-SLC12A8-1 group vs. si-con group was 209.67 ± 24.58 vs. 339.33 ± 27.5 (p < 0.01, Supplementary Fig. B-C).
The effect of loss- and gain-of-function of SLC12A8 on the breast carcinoma cells invasion and migration. Transwell assays were implemented to examine the invasive and migratory abilities of MCF7 cells with knockdown of SLC12A8 (a, b) and Bcap-37 cells with overexpression of SLC12A8 (c, d). a, c Representative photographs of invasion and migration cells. b, d The number of invaded and migrated cells was quantified. **p < 0.01. Magnification, ×200
On the other hand, enhanced expression of SLC12A8 in Bcap-37 cells presented an opposite outcome, as we observed that the number of invaded and migrated cells was obvious more than their respective vector group (p < 0.01, invasion: 199.33 ± 12.66 vs. 111.33 ± 8.33; migration: 268 ± 20.52 vs. 144.33 ± 8.02 Fig. 3c, d). Collectively, these results illustrated that SLC12A8 could enhance the invasiveness and motility of breast carcinoma cells.
SLC12A8 is closely linked to Toll-like receptor/NOD like receptor (TLR/NLR) signaling pathway
To explore the potential mechanism of the promoting-effect of SLC12A8 in breast carcinoma, GSEA was conducted according to the SLC12A8 expression profile from TCGA database. We observed that the gene sets of “KEGG_TOLL_LIKE_RECEPTOR” and “KEGG_NOD_LIKE_RECEPTOR” were significantly enriched in patients with high SLC12A8 expression (p < 0.01, Figs. 4a and 5a). Then we firstly examined the effect of SLC12A8 on the TLR signaling pathway-related biomarkers TLR4, MyD88, p-NF-κB, NF-κB, p-IκBα, and IκBα by western blot analysis. SLC12A8-2-depleted MCF7 cells showed less protein levels of TLR4, MyD88, p-NF-κB, and p-IκBα in comparison with their relative si-con group (p < 0.01, Fig. 4b, c, f, g). However, no obvious change was observed on the level of NF-κB and IκBα after knockdown of SLC12A8 compared with control (p > 0.05, Fig. 4b, c, f, g). Conversely, overexpression of SLC12A8 in Bcap-37 cells led to a significant increase on the protein levels of TLR4, MyD88, p-NF-κB, and p-IκBα in comparison with relative vector group (p < 0.01, Fig. 4d, e, f, g). But no difference was observed in NF-κB and IκBα protein expression (p > 0.05, Fig. 4d, e, f, g). All these data suggested that the promoting effect of SLC12A8 in breast carcinoma may be related to the activation of TLR pathway.
Correlation between SLC12A8 and NOD_LIKE_RECEPTOR signaling pathway-related markers. GSEA revealed that the NOD_LIKE_RECEPTOR pathway (a) was enriched in the high SLC12A8 expression group according to the TCGA database. Protein levels of NOD1, NOD2, NLRP3, and IL-1β in MCF7 (b, d) and Bcap-37 (c, e) cells were determined by western blot analysis after altering the expression of SLC12A8. **p < 0.01
Correlation between SLC12A8 and TOLL_LIKE_RECEPTOR signaling pathway-related markers. GSEA revealed that the TOLL_LIKE_RECEPTOR pathway (a) was enriched in the high SLC12A8 expression group according to the TCGA database. b–e Protein levels of TLR4, MyD88, p-NF-κB, and NF-κB in MCF7 (b, c) and Bcap-37 (d, e) cells were determined by western blot analysis after altering the expression of SLC12A8. f, g Protein levels of p-IκBα, IκBα, and pro-caspase-1 were determined by western blot analysis after altering SLC12A8 expression. *p < 0.05, **p < 0.01
Next, western blot analysis revealed a positive correlation between SLC12A8 expression and NLR signaling pathway-related biomarkers, as we observed that silencing SLC12A8 led to significant reductions on the protein levels of NOD1, NOD2, NLRP3, pro-caspase-1 and IL-1β, whereas overexpression of SLC12A8 resulted in obvious elevations on the levels of these biomarkers (p < 0.01, Figs. 4f, g and 5b–e). So we inferred that SLC12A8 play the cancerogenic role in breast carcinoma partly by activating the TLR/NLR signaling.
Discussion
Cancer is a preventable disease if detected at an early stage (Banik et al. 2020). Advances in whole-genome sequencing and bioinformatics have indeed opened new directions for cancer research, but they are not enough. Breast carcinoma is highly heterogeneous at the molecular level, and the exploration of key molecules that regulate breast carcinoma progression would provide valuable targets for the diagnosis and therapeutic intervention of breast carcinoma (Agosto-Arroyo et al. 2014). Thus, our work focuses on the investigation of the role of SLC12A8 in the diagnosis and prognosis of the breast carcinoma, and evaluation of its biological roles, as well as the potential mechanisms, hoping to provide a possible bio-target for the therapy of breast carcinoma.
Members of SLC12 family are the most commonly used targets for diuretics and have been shown to be disease-caused genes for genetic disorders, and may play a role in multi-gene complex diseases such as arterial hypertension, epilepsy and cancer (Arroyo et al. 2013). The role of these members in breast carcinoma is gradually being explored and reported. It has been shown that overexpression of SCL12A6 and SCL12A7 could significantly promote the MCF7 cells proliferation and invasion (Hsu et al. 2007). Besides, SCL12A7 was found to be overexpressed in breast carcinoma (Brown et al. 2018). Importantly, a study showed that SLC12A8 was associated with the overall survival in patients with breast carcinoma (Kim et al. 2018), inspiring us to systematically investigate its biological functions and the underlying mechanisms. Consistently, in our work, SLC12A8 was found to be highly expressed in patients with breast carcinoma according to the public datasets and showed an independent prognostic value. All these data support that SLC12A8 may be a useful marker for the diagnosis and prognosis of breast carcinoma. Moreover, functional experiments in vitro suggested that SLC12A8 could significantly enhance the viability, invasiveness and motility of breast carcinoma cells. However, the function of this protein has not been clearly defined (Arroyo et al. 2013). A reasonable explanation is that a splicing variant SLC12A8 affects the transport of polyamines, which play an important role in cell death and proliferation by maintaining the high concentration in cancer cells (Daigle et al. 2009; Linsalata et al. 2010).
Numerous studies have shown that the occurrence of many cancers is related to the dysfunction of signaling pathways (Soysal et al. 2015). Identifying relevant signaling pathways can help identify targeted drugs for the treatment of breast carcinoma (Sliva 2004). Thus, to determine the SLC12A8-related signaling pathways involved in breast carcinoma, we found that high expression of SLC12A8 had positive correlations with the pattern recognition receptors TLR and NLR by GSEA. These data raise a question that whether the growth- and motion-promoting effects of SLC12A8 in breast carcinoma are linked with the TLR and NLR signaling pathways-related proteins. TLRs play an important role in the immune system, which can be expressed in both immune cells and tumor cells (Green et al. 2014). According to whether the downstream signal transduction needs MyD88, the TLR signaling pathway can be divided into MyD88-dependent pathway and MyD88-independent pathway (Takeda and Akira 2004). An important character of the MyD88-dependent pathway is the early activation of NF-κB (Youn et al. 2008). It was shown that TLR4 is associated with the occurrence and distant metastasis in breast carcinoma (Chen et al. 2015; Yang et al. 2014). Moreover, Ma et al. clarified that TLR4-MyD88/MyD88 is a prognostic risk factor for breast carcinoma (Ma et al. 2014). All these studies suggest that inhibition of TLR4 and MyD88 expression and suppression of NF-κB phosphorylation may be beneficial in preventing the development of breast carcinoma. Coincidently, our experiment achieved the expected results, as observed that SLC12A8 presents a positively correlation with the protein expression of TLR4, MyD88, p-NF-κB and p-IκBα. However, the expression of NF-κB and IκBα does not change with the expression of SLC12A8. Therefore, we inferred that SLC12A8 promotes cells proliferation, invasion and migration partly by activating the TLR signaling pathway.
Inflammation is closely related to tumors and is even a major driver of many tumors (Korniluk et al. 2017). NOD, as one of the representatives of inflammatory immune receptors, is worth exploring in relationship with breast carcinoma. NODs and NOD like protein 3 (NLRP3) are a group of evolutionarily conserved intracellular pattern recognition receptors (Zhong et al. 2013). NOD1 and NOD2 activates NF-κB, MAPK, JNK, p38 and ERK signaling pathways and stimulates the expression of various inflammatory factors, such as TNF-α, IL-1β, IL-6, IL-8 and IL-12 (Lappas 2013; Ogura et al. 2001). NLRP3 inflammasome activates Caspase1 and mediates the release of IL-18 and IL-1β (Dai et al. 2017). Thus, to evaluate the correlation between SLC12A8 and NLR signaling pathway, we detected the protein expression changes of NOD1, NOD2, NLRP3, pro-caspase-1 and IL-1β after altering SLC12A8 expression. The results showed that SLC12A8 had a positively correlation with these markers. Therefore, it is reasonable to speculate that SLC12A8 may affect the biological phenotypes in breast carcinoma by activating the TLR/NLR signaling pathway.
Taken together, the current study revealed that SLC12A8 has a prognostic value in breast carcinoma, and suggested that during the progression of breast carcinoma, SLC12A8 promoted the cells proliferation, migration, and invasion partly through regulating the TLR/NLR signaling pathway. These results highlight the potential of SLC12A8 to be a target for the therapeutic intervention of breast carcinoma.
References
Agosto-Arroyo E, Isayeva T, Almeida JS, Harada S (2014) Molecular profiling of breast ductal carcinoma in-situ. Pathology 46:S29
Arroyo JP, Kahle KT, Gamba G (2013) The slc12 family of electroneutral cation-coupled chloride cotransporters. Mol Aspects Med 34:288–298
Banik K, Ranaware AM, Harsha C, Nitesh T, Girisa S, Deshpande V, Fan L, Nalawade SP, Sethi G, Kunnumakkara AB (2020) Piceatannol: a natural stilbene for the prevention and treatment of cancer. Pharmacol Res 153:104635
Brown TC, Murtha TD, Rubinstein JC, Korah R, Carling T (2018) Slc12a7 alters adrenocortical carcinoma cell adhesion properties to promote an aggressive invasive behavior. Cell Commun Signal CCS 16:27
Chen X, Zhao F, Zhang H, Zhu Y, Wu K, Tan G (2015) Significance of tlr4/myd88 expression in breast cancer. Int J Clin Exp Pathol 8:7034–7039
Dai X, Tohyama M, Murakami M, Sayama K (2017) Epidermal keratinocytes sense dsrna via the nlrp3 inflammasome, mediating interleukin (il)-1beta and il-18 release. Exp Dermatol 26:904–911
Daigle ND, Carpentier GA, Frenette-Cotton R, Simard MG, Lefoll MH, Noel M, Caron L, Noel J, Isenring P (2009) Molecular characterization of a human cation-cl- cotransporter (slc12a8a, ccc9a) that promotes polyamine and amino acid transport. J Cell Physiol 220:680–689
Feigin ME, Garvin T, Bailey P, Waddell N, Chang DK, Kelley DR, Shuai S, Stein LD, Biankin AV, Schatz MC, Tuveson DA, Gallinger S, McPherson JD, Grimmond SM, Khurana E (2017) Recurrent noncoding regulatory mutations in pancreatic ductal adenocarcinoma. Nat Genet 49:825–833
Gagnon KB, Delpire E (2013) Physiology of slc12 transporters: Lessons from inherited human genetic mutations and genetically engineered mouse knockouts. Am J Physiol Cell Physiol 304:C693–C714
Green TL, Santos MF, Ejaeidi AA, Craft BS, Lewis RE, Cruse JM (2014) Toll-like receptor (tlr) expression of immune system cells from metastatic breast cancer patients with circulating tumor cells. Exp Mol Pathol 97:44–48
Hebert SC, Mount DB, Gamba G (2004) Molecular physiology of cation-coupled cl- cotransport: the slc12 family. Pflugers Arch 447:580–593
Hsu YM, Chou CY, Chen HH, Lee WY, Chen YF, Lin PW, Alper SL, Ellory JC, Shen MR (2007) Igf-1 upregulates electroneutral k-cl cotransporter kcc3 and kcc4 which are differentially required for breast cancer cell proliferation and invasiveness. J Cell Physiol 210:626–636
Kim JE, Choi J, Park J, Park C, Lee SM, Park SE, Song N, Chung S, Sung H, Han W, Lee JW, Park SK, Kim MK, Noh DY, Yoo KY, Kang D, Choi JY (2018) Associations between genetic polymorphisms of membrane transporter genes and prognosis after chemotherapy: meta-analysis and finding from seoul breast cancer study (sebcs). Pharmacogenom J 18:633–645
Korniluk A, Koper O, Kemona H, Dymicka-Piekarska V (2017) From inflammation to cancer. Ir J Med Sci 186:57–62
Kozlowski J, Kozlowska A, Kocki J (2015) Breast cancer metastasis - insight into selected molecular mechanisms of the phenomenon. Postep Higieny i Med Dosw (Online) 69:447–451
Lappas M (2013) Nod1 and nod2 regulate proinflammatory and prolabor mediators in human fetal membranes and myometrium via nuclear factor-kappa b. Biol Reprod 89:14
Linsalata M, Notarnicola M, Tutino V, Bifulco M, Santoro A, Laezza C, Messa C, Orlando A, Caruso MG (2010) Effects of anandamide on polyamine levels and cell growth in human colon cancer cells. Anticancer Res 30:2583–2589
Ma FJ, Liu ZB, Hu X, Ling H, Li S, Wu J, Shao ZM (2014) Prognostic value of myeloid differentiation primary response 88 and toll-like receptor 4 in breast cancer patients. PLoS One 9:e111639
Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G (2001) Nod2, a nod1/apaf-1 family member that is restricted to monocytes and activates nf-kappab. J Biol Chem 276:4812–4818
Siegel RL, Miller KD, Sauer AG, Fedewa SA, Jemal A (2020) Cancer statistics, 2020. CA A Cancer J Clin 70:7–30
Sliva D (2004) Signaling pathways responsible for cancer cell invasion as targets for cancer therapy. Curr Cancer Drug Targets 4:327–336
Soysal SD, Tzankov A, Muenst SE (2015) Role of the tumor microenvironment in breast cancer. Pathobiology 82:142–152
Takeda K, Akira S (2004) Tlr signaling pathways. Semin Immunol 16:3–9
Tao Z, Shi A, Lu C, Song T, Zhang Z, Zhao J (2015) Breast cancer: epidemiology and etiology. Cell Biochem Biophys 72:333–338
Teng F, Guo M, Liu F, Wang C, Dong J, Zhang L, Zou Y, Chen R, Sun K, Fu H, Fu Z, Guo W, Ding G (2016) Treatment with an slc12a1 antagonist inhibits tumorigenesis in a subset of hepatocellular carcinomas. Oncotarget 7:53571–53582
Weigelt B, Peterse JL, van ’t Veer LJ (2005) Breast cancer metastasis: markers and models. Nat Rev Cancer 5:591–602
Xu L, Li X, Cai M, Chen J, Li X, Wu WK, Kang W, Tong J, To KF, Guan XY, Sung JJ, Chan FK, Yu J (2016) Increased expression of solute carrier family 12 member 5 via gene amplification contributes to tumour progression and metastasis and associates with poor survival in colorectal cancer. Gut 65:635–646
Yang H, Wang B, Wang T, Xu L, He C, Wen H, Yan J, Su H, Zhu X (2014) Toll-like receptor 4 prompts human breast cancer cells invasiveness via lipopolysaccharide stimulation and is overexpressed in patients with lymph node metastasis. PLoS One 9:e109980
Youn HS, Lim HJ, Choi YJ, Lee JY, Lee MY, Ryu JH (2008) Selenium suppresses the activation of transcription factor nf-kappa b and irf3 induced by tlr3 or tlr4 agonists. Int Immunopharmacol 8:495–501
Zhang Y, Liu Y, Ye Y, Shen D, Zhang H, Huang H, Li S, Wang S, Ren J (2017) Quantitative proteome analysis of colorectal cancer-related differential proteins. J Cancer Res Clin Oncol 143:233–241
Zhong Y, Kinio A, Saleh M (2013) Functions of nod-like receptors in human diseases. Front Immunol 4:333
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10616_2020_439_MOESM1_ESM.tif
Supplementary Figure The effects of si-SLC12A8-1 on the breast carcinoma cells proliferation, invasion and migration.(A) After transfected with si-con and si-SLC12A8-1 in MCF7 cells, the optical density values were determined by CCK8 assays. (B-C) Transwell assays were used to examine the invasive and migratory abilities of MCF7 cells after transfected with si-con and si-SLC12A8-1. (B) Representative photographs of invasion and migration cells. (C) The number of invaded and migrated cells was quantified. Magnification, x200. **p<0.01. Supplementary file1 (TIFF 2680 kb)
Rights and permissions
About this article
Cite this article
Li, L., Xia, J., Cui, R. et al. Solute carrier family 12 member 8 impacts the biological behaviors of breast carcinoma cells by activating TLR/NLR signaling pathway. Cytotechnology 73, 23–34 (2021). https://doi.org/10.1007/s10616-020-00439-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10616-020-00439-y