Abstract
Mouse embryonic stem (ES) cells and induced pluripotent stem (iPS) cells have the ability to differentiate in vitro into various cell lineages including neurons. The differentiation of these cells into neurons has potential applications in regenerative medicine. Previously, we reported that a chick dorsal root ganglion (DRG)-conditioned medium (CM) promoted the differentiation of mouse ES and iPS cells into neurons. Here, we used real-time PCR to investigate the differentiation patterns of ES and iPS cells into neurons when DRG-CM was added. DRG-CM promoted the expression levels of βIII-tubulin gene (a marker of postmitotic neurons) in ES and iPS cells. ES cells differentiated into neurons faster than iPS cells, and the maximum peaks of gene expression involved in motor, sensory, and dopaminergic neurons were different. Rho kinase (ROCK) inhibitors could be very valuable at numerous stages in the production and use of stem cells in basic research and eventual cell-based therapies. Thus, we investigated whether the addition of a ROCK inhibitor Y-27632 and DRG-CM on the basis of the differentiation patterns promotes the neuronal differentiation of ES cells. When the ROCK inhibitor was added to the culture medium at the initial stages of cultivation, it stimulated the neuronal differentiation of ES cells more strongly than that stimulated by DRG-CM. Moreover, the combination of the ROCK inhibitor and DRG-CM promoted the neuronal differentiation of ES cells when the ROCK inhibitor was added to the culture medium at day 3. The ROCK inhibitor may be useful for promoting neuronal differentiation of ES cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mouse embryonic stem (ES) cells have been derived from the inner cell mass of 3.5-day-old blastocysts of preimplantation mouse embryos. On the other hand, mouse induced pluripotent stem (iPS) cells have been directly generated from mouse embryonic fibroblasts by introducing the four transcription genes Oct3/4, Sox2, c-Myc, and Klf4. Both ES and iPS cells are pluripotent and can differentiate in vitro into various cell lineages including neurons (Evans and Kaufman 1981; Martin 1981; Takahashi and Yamanaka 2006). iPS cells may be used as patient-specific pluripotent stem cells for studies of disease pathogenesis, drug discovery, and transplantation therapy. Therefore, research efforts have been focused on controlling the differentiation of ES and iPS cells into neurons to understand their potential applications in neuroscience and regenerative medicine (Kitazawa and Shimizu 2011).
Previously, we reported that a chick dorsal root ganglion (DRG)-conditioned medium (CM) promoted the differentiation of mouse ES and iPS cells into neurons at 11–12 days of cultivation (Kitazawa and Shimizu 2005, 2011). In addition, we demonstrated that the percentage of neurons differentiated from mouse ES cells was approximately 50 %. A total of 40–60 % neurons that differentiated from ES cells were primarily motor neurons (Kitazawa and Shimizu 2007). Because ES and iPS cells may produce a variety of specialized cell types, we require to control and manipulate cell differentiation to produce exclusive populations of specific cell types. Therefore, we need to clarify the process of formation of the cells differentiated from ES and iPS cells.
Rho kinase (ROCK) plays key roles in mediating the control of the actin cytoskeleton through Rho family GTPases in response to extracellular signals. Such signaling pathways contribute to diverse neuronal functions such as cell migration, axonal guidance, dendritic spine morphology, axonal regeneration, and cell survival (Schmandke et al. 2007). Because ROCK regulates the activities of many target proteins by its kinase activity, the inhibition of ROCK activity may provide new perspectives for controlling the in vitro differentiation of ES and iPS cells into neurons. Several researchers reported the effects of the ROCK inhibitor on the neuronal differentiation of bone marrow-derived mesenchymal stem cells, mouse ES cells, adipose tissue-derived stem cells, and mouse neural stem cells (Pacary et al. 2006; Chang et al. 2011; Kamishibahara et al. 2014; Xue et al. 2012; Gu et al. 2013). Minase et al. (2010) reported that the ROCK inhibitor potentiated nerve growth factor (NGF)-induced neurite outgrowth in PC12 cells. In this study, we investigated the differentiation patterns of ES and iPS cells into neurons using real-time PCR. To achieve the efficient differentiation of ES cells into neurons on the basis of the differentiation patterns, we also investigated the effects of the combination of the ROCK inhibitor Y-27632 and DRG-CM on the differentiation of ES cells.
Materials and methods
Cultivation and colony formation of mouse ES and iPS cells
Mouse ES cells (129SV; Dainippon Pharmaceutical, Osaka, Japan) after 16–20 passages were grown on a mitotically inactivated mouse embryonic fibroblast (PMEF-H-C; Millipore, Temecula, CA, USA) feeder layer in DMEM (SLM-220-B; Millipore) supplemented with 15 % knockout serum replacement (10828-028; Gibco BRL, Grand Island, NY, USA), 1 % nucleosides (ES-008-D; Millipore), 1 mM non-essential amino acids (TMS-001-C; Millipore), 0.1 mM 2-mercaptoethanol (ES-007-E; Millipore), 1 % l-glutamine (TMS-002-C; Millipore), 1 % penicillin/streptomycin (15140-122; Gibco BRL), and 0.1 % leukemia inhibitory factor (125-05603; Wako Pure Chemical Industries, Osaka, Japan) at 37 °C in a humidified atmosphere of 5 % CO2 using 0.1 % gelatin (521-00325; Wako Pure Chemical Industries, Osaka, Japan)-coated 100-mm culture dishes (3020-100; Iwaki, Tokyo, Japan). Mouse iPS cells (iPS-MEF-Ng-20D-17; APS0001; Riken Cell Bank, Saitama, Japan) were grown as described previously (Kitazawa and Shimizu 2011). To form spherical ES cell or iPS cell colonies, approximately 4 × 105 cells were plated on a non-adhesive 100-mm plastic dish (AU2010; Eikenkizai, Tokyo, Japan) with DMEM and were cultured for 6–9 days at 37 °C in a humidified atmosphere of 5 % CO2. Half of the medium was replaced with a fresh medium every 3 days.
Preparation of DRG-CM
DRGs were dissected from 8-day-old chick embryos as described previously (Naka et al. 2002). Sixty DRGs per dish were plated on gelatin-coated 100-mm culture dishes with a DMEM/F-12 K medium containing 10 ng/ml of NGF (2256X; Techne, Minneapolis, MN, USA) and were cultured for 2 days at 37 °C in a humidified atmosphere of 5 % CO2. The DMEM/F-12 K medium consisted of 49 % DMEM (R-SLM-220-B; Dainippon Pharmaceutical) and 49 % F-12 nutrient mixture (21127-022; Gibco BRL), which contained 1 % N-2 supplement (17502-048; Gibco BRL) instead of serum and 1 % penicillin/streptomycin. The supernatant of the culture medium derived by centrifugation was filtered through a 0.22-µm filter (SLGV033RS, Millipore, Bedford, MA, USA); this filtrate was designated as DRG-CM.
Differentiation of mouse ES and iPS cells
To investigate the effects of chick DRG-CM on ES cell or iPS cell differentiation, the spherical and undifferentiated colonies (approximately 200 µm in diameter) were taken out of the non-adhesive 100-mm plastic dishes using a 200-µl siliconized pipette tip attached to a sterile pipette, and one colony per well was plated with the DMEM/F-12 K medium on a gelatin-coated 96-well assay plate (353948; Becton–Dickinson, Franklin Lakes, NJ, USA). After 5 % DRG-CM was added to 7–15 replicate wells, colonies were cultured for 3–18 days at 37 °C in a humidified atmosphere of 5 % CO2. Half of the medium was replaced with a fresh medium containing DRG-CM every 3 days.
Gene expression analysis by real-time PCR
The extraction of total RNA was performed using an RNeasy Mini Kit (74104; Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The potentially contaminating genomic DNA was digested by RNase-free DNase set (79254; Qiagen) for 15 min at room temperature. cDNA was synthesized using a High Capacity cDNA Reverse Transcription Kit (4368814; Applied Biosystems, Carlsbad, CA, USA). Real-time PCR was performed using a KAPA SYBR FAST qPCR Kit Master Mix (2X) ABI Prism (KR0390; Kapa Biosystems, Boston, MA, USA) and Step One Real-Time PCR system (Applied Biosystems). The cycling conditions were as follows: 3 min of pre-denaturation at 95 °C followed by 40 cycles of 30-s denaturation at 95 °C, 30 s at 60 °C, and 30 s at 75 °C. The standard serial five-fold dilutions of template cDNA were repeated on every plate. The following sense and antisense primers were designed: nestin (Nakayama et al. 2003), 5′-CTCGAGCAGGAAGTGGTAGG-3′ and 5′-TTGGGACCAG GGACTGTTAG-3′; βIII-tubulin, 5′-AGGCCCGACAACTTTATCTTTGGTC-3′ and 5′-TGCAGTAGGTC TCGTCTGTGTTCTC-3′; Lim-3, 5′-TTAAGCGCTTCGGGACCAAG-3′ and 5′-TCCTGGATGCTGTCC TTGTCG-3′; Brn-3, 5′-GCCTCACTTTGCCATGCATC-3′ and 5′-CAGGGCACGCTATTCATCGT-3′; tyrosine hydroxylase (TH) (Nakayama et al. 2003), 5′-AATTCCCCACGTGGAATACA-3′ and 5′-CTGCT GTGTCTGGGTCAAAG-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Nakayama et al. 2003), 5′-ACTCACGGCAAATTCAACGG-3′ and 5′-ACGTCAGATCCACGACGGAC-3′. The expression levels were normalized against GAPDH.
Measurement of the number of types of neurons
Differentiated ES cell colonies cultivated on a gelatin-coated 96-well culture plates were washed three times with cold phosphate-buffered saline (PBS). The colonies were incubated with a trypsin/EDTA solution (SM-2003-C; Millipore) for 2 min at room temperature and were subsequently incubated in DMEM supplemented with 15 % knockout serum replacement, 1 % nucleosides, 1 mM non-essential amino acids, 0.1 mM 2-mercaptoethanol, 1 % l-glutamine, and 1 % penicillin/streptomycin. After removing the supernatant by centrifugation, we washed the colonies with cold PBS. The cells were dispersed by pipetting, and approximately 1 × 106 cells were transferred to a 1.5 ml tube. The cells were then washed with cold PBS and were incubated with 4 % paraformaldehyde phosphate buffer solution for 60 min on ice. We washed the cells with cold PBS after removing the supernatant by centrifugation. The cells were then incubated with a cold 99.8 % methanol solution for 30 min at −80 °C. After two washes with cold PBS, the cells were incubated with primary antibodies overnight at 4 °C. The following primary antibodies were used for labeling: anti-βIII-tubulin (MAB1637; Millipore), anti-Lim-3 (AB3202; Millipore), anti-Brn-3 (SC6026; Santa Cruz Biotechnology, Santa Cruz, CA, USA), and anti-TH (2792S; Cell Signaling Technology, Danvers, MA, USA) antibodies. After three washes with cold PBS, the cells were incubated for 60 min at room temperature with an Alexa Fluor 488-labeled secondary antibody (A11055 or A11008; Molecular Probes, Eugene, OR, USA). Following another three cold PBS washes, the cells were incubated with Hoechst 33258 (17528; ABD Bioquest, Sunnyvale, CA, USA) for 30 min at room temperature to visualize the nuclei. The cells were then washed two times with PBS, and we measured the number of fluorescence-activated cells using seven randomly chosen immunofluorescence images using a confocal microscope (IX81; Olympus, Tokyo, Japan), a confocal scanning unit (CSU-X1; Yokogawa Electric, Tokyo, Japan), and a CCD camera (iXon DU897; Andor Technology, Belfast, UK).
Immunofluorescence analysis
Differentiated ES cell colonies were cultivated in a gelatin-coated 96-well assay plate, washed three times with cold PBS, and fixed with 4 % paraformaldehyde phosphate buffer solution for 30 min at room temperature. After three PBS washes, the cells were incubated with cold 99.8 % methanol for 15 min at −80 °C. The cells were then washed three times with cold PBS and were blocked with a 5 % bovine serum albumin solution containing 0.3 % Triton X-100 for 1 h at room temperature or overnight at 4 °C. Next, the cells were incubated with primary antibodies at 4 °C overnight, washed three times with PBS, and then incubated with the secondary antibody for 60 min at room temperature. The cells were subsequently washed three times with PBS, and incubated with Hoechst 33258 for 30 min at room temperature for nuclear staining. After washing two times with PBS, we measured the fluorescence intensity of the cells using a fluoro-image analyzer (FLA-3000R; Fujifilm, Tokyo, Japan).
Statistical analysis
Data were calculated as mean ± standard error of the mean (SEM). Statistical comparisons were made using one-way ANOVA and the Tukey–Kramer multiple comparison post hoc test. Values of P less than 0.05 were considered statistically significant.
Results
Gene expression analysis during the differentiation processes of ES and iPS cells by real-time PCR
We investigated the differentiation processes of ES and iPS cells into neurons using real-time PCR when 5 % DRG-CM was added to the culture medium. Nestin (a marker of neural stem cells) and βIII-tubulin (a marker of postmitotic neurons) were examined as genetic markers in differentiated ES and iPS cells using real-time PCR as shown in Fig. 1. Figure 1a, b show that the expression levels of nestin gene increased in both ES and iPS cells until day 9 and decreased thereafter. The addition of DRG-CM did not promote the expression levels of nestin gene in both ES and iPS cells. Figure 1c, d show that the expression levels of βIII-tubulin gene in ES and iPS cells were promoted by the addition of DRG-CM. The highest expression levels of βIII-tubulin gene in ES and iPS cells were attained at days 9 and 15, respectively. Moreover, ES cells differentiated into neurons faster than iPS cells.
Expression patterns of nestin and βIII-tubulin genes in ES and iPS cells using real-time PCR. a Expression patterns of nestin (a marker of neural stem cells) gene in ES cells. b Expression patterns of nestin gene in iPS cells. c Expression patterns of βIII-tubulin (a marker of postmitotic neurons) gene in ES cells. d Expression patterns of βIII-tubulin gene in iPS cells. Data are presented as mean ± SEM of three replicates. Open bars ES and iPS cell colonies cultured without DRG-CM. Solid bars ES and iPS cell colonies cultured with 5 % DRG-CM
We investigated the gene expression patterns of Lim-3 (a marker of cranial motor neurons), Brn-3 (a marker of sensory neurons), and TH (a marker of dopaminergic neurons) in ES and iPS cells as shown in Fig. 2. Because ES and iPS cells predominantly differentiated into motor and sensory neurons (Kitazawa and Shimizu 2011). The highest expression levels of Lim-3, Brn-3, and TH genes in ES and iPS cells were attained at day 12 (Fig. 2a, b), days 15 and 18 (Fig. 2c, d), and days 9 and 15 (Fig. 2e, f), respectively. These results show that the rate of differentiation into each cell type was different.
Expression patterns of Lim-3, Brn-3, and TH genes in ES and iPS cells using real-time PCR. a Expression patterns of Lim-3 (a marker of cranial motor neurons) gene in ES cells. b Expression patterns of Lim-3 gene in iPS cells. c Expression patterns of Brn-3 (a marker of sensory neurons) gene in ES cells. d Expression patterns of Brn-3 gene in iPS cells. e Expression patterns of TH (a marker of dopaminergic neurons) gene in ES cells. f Expression patterns of TH gene in iPS cells. Data are presented as mean ± SEM of three replicates. Open bars ES and iPS cell colonies cultured without DRG-CM. Solid bars ES and iPS cell colonies cultured with 5 % DRG-CM
Number of types of neurons
We measured the number of types of neurons differentiated from ES cells using a confocal laser scanning microscope. Figure 3 shows the numbers of motor neurons labeled with the anti-Lim-3 antibody, sensory neurons labeled with the anti-Brn-3 antibody, and dopaminergic neurons labeled with the anti-TH antibody at days 12, 15, and 18. The population of motor neurons was composed of 19, 12.5, and 10 % of the total cells at days 12, 15, and 18, respectively. The rate of motor neuron differentiation decreased from day 15 in a similar manner to the gene expression pattern of motor neurons (Lim-3). Moreover, the numbers of sensory and dopaminergic neurons were similar to their gene expression patterns.
Percentage of motor, sensory, and dopaminergic neurons differentiated from ES cells. ES cell colonies were cultured for 12, 15, and 18 days with 5 % DRG-CM. The number of fluorescent cells was measured using a confocal laser scanning microscope. Lim-3, a marker of cranial motor neurons; Brn-3, a marker of sensory neurons; TH, a marker of dopaminergic neurons. Data are presented as mean ± SEM of seven replicates
Effects of addition of the ROCK inhibitor on differentiation of ES cells into neurons
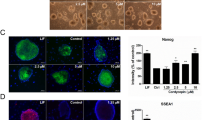
We investigated whether the addition of 20 µM of the ROCK inhibitor Y-27632 (253-00513; Wako Pure Chemical Industries) together with 5 % DRG-CM at the initial culture stages promotes the neuronal differentiation of ES cells by synergistic effects. Figure 4 shows the neuronal differentiation of ES cells in the presence of the ROCK inhibitor. When the ROCK inhibitor alone or in combination with DRG-CM was added to the culture medium at days 0 or 3, the neuronal differentiation of ES cells was promoted. However, the ROCK inhibitor did not promote the neuronal differentiation of ES cells when it was added to the culture medium after day 6. Notably, the ROCK inhibitor stimulated the neuronal differentiation of ES cells more strongly than that stimulated by DRG-CM without synergistic effects. Figure 5 shows the fluorescence micrographs of differentiated ES cell colonies with neurite outgrowth at day 12. ES cell colonies clearly exhibited neurite outgrowth after addition of either the ROCK inhibitor alone or in combination with the DRG-CM at day 3. These results are in line with the results of the immunofluorescence analysis.
Effects of the ROCK inhibitor on neuronal differentiation of ES cells. ES cell colonies were cultured for 12, 15, and 18 days. a Fluorescence intensity of neurons labeled with an antibody against βIII-tubulin. b Fluorescence intensity of motor neurons labeled with an antibody against Lim-3. c Fluorescence intensity of sensory neurons labeled with an antibody against Brn-3. d Fluorescence intensity of dopaminergic neurons labeled with an antibody against TH. DRG-CM, differentiated ES cell colonies cultured with DRG-CM; ROCK inhibitor, differentiated ES cell colonies cultured with the ROCK inhibitor; DRG-CM + ROCK inhibitor at day 3, differentiated ES cell colonies cultured with DRG-CM and the ROCK inhibitor added at day 3; DRG-CM + ROCK inhibitor at day 6, differentiated ES cell colonies cultured with DRG-CM and the ROCK inhibitor added at day 6; DRG-CM + ROCK inhibitor at day 9, differentiated ES cell colonies cultured with DRG-CM and the ROCK inhibitor added at day 9. The fluorescence intensity of neurons differentiated from ES cells with DRG-CM addition was represented as 100 %. Data are presented as mean ± SEM of seven replicates. *P < 0.05 and **P < 0.01 compared with DRG-CM
Discussion
We investigated the differentiation patterns of ES and iPS cells into neurons using real-time PCR to clarify the differentiation stages when 5 % DRG-CM was added to the culture medium. With real-time PCR, large numbers of samples can be rapidly tested using small amounts of RNA, and accurate results can be obtained. The expression levels of nestin gene in both ES and iPS cells increased until day 9 and decreased thereafter. The addition of DRG-CM did not promote the expression levels of nestin gene, but it did promote the expression levels of βIII-tubulin gene in both ES and iPS cells. The highest expression levels of βIII-tubulin gene in ES and iPS cells were attained at days 9 and 15, respectively, followed by a gradual decrease (Fig. 1). Similarly Xie et al. (2009) reported that the expression levels of smooth muscle cell-specific genes gradually decreased after the peak of the gene expression levels. Furthermore, we report that ES cells differentiated into neurons faster than iPS cells, in agreement with a study by Morizane et al. (2009), which reported that the differentiation of iPS cells into renal lineages was slower than that of ES cells. We also found that the maximum peaks of gene expression involved in motor, sensory, and dopaminergic neurons were different (Fig. 2). The differentiation rates of individual neurons appeared to be dependent on the types of neurons. The TH gene expression involved in dopaminergic neurons in ES cells was faster than the Lim-3 and Brn-3 gene expression involved in motor and sensory neurons, respectively.
To obtain the desired type of neuron, the maximum period of differentiation for a particular neuron must be considered. The levels of gene expression decreased after they reached the maximum level. This indicates that the amount of the desired mRNA detected in the cells decreased and undifferentiated cells proliferated during differentiation (Noaksson et al. 2005). Because the gene expression levels of differentiated cells do not indicate the population, we investigated the number of the differentiated cells using immunofluorescence analysis (Fig. 3). We found that the percentage of motor neurons was 19 % at day 12. This value was similar to that of a previous study (Kitazawa and Shimizu 2007). Because the population of neurons was 52 % at day 11 (Kitazawa and Shimizu 2007), neurons differentiated from ES cells were predominantly motor neurons. Then, the percentages of motor neurons gradually decreased in a similar manner to the changes of the gene expression levels. The percentages of sensory and dopaminergic neurons were lower than that of motor neurons. In addition, we demonstrated that ES cells differentiated into muscle cells other than neurons. The percentages of muscle cells increased to 64 % after 15 days (data not shown). Wobus et al. (1988) also reported that mouse ES cells differentiated into muscle cells slower than neurons.
ROCK is one of the major downstream mediators of Rho, which plays crucial regulatory roles in cellular proliferation and differentiation. ROCK is involved in a wide range of fundamental cellular functions such as contraction, adhesion, migration, and proliferation. Furthermore, ROCK plays an important role in the regulation of apoptosis in various cell types and animal disease models. Therefore, ROCK inhibitors could be very valuable at numerous stages in the production and use (culturing, genetic modification, differentiation, and implantation) of stem cells in basic neuroscience research and eventual cell-based therapies (Shi and Wei 2007; Olson 2008). Several researchers reported the promotion of neuronal differentiation of various stem cells by the addition of the ROCK inhibitor (Pacary et al. 2006; Chang et al. 2011; Xue et al. 2012; Gu et al. 2013). In addition, we reported that the ROCK inhibitor has a significant potential to regulate the differentiation of the ES cells (Kamishibahara et al. 2014). Therefore, we investigated whether the combination of the ROCK inhibitor Y-27632 and DRG-CM promoted the neuronal differentiation of ES cells by synergistic effects (Fig. 4). The ROCK inhibitor together with DRG-CM was added to the culture medium at days 3, 6, and 9 on the basis of the differentiation patterns. Then, ES cell colonies were cultured for 12–18 days. We found that the ROCK inhibitor promoted the neuronal differentiation of ES cells more strongly than that promoted by DRG-CM. The ES cell colonies stimulated by the ROCK inhibitor at the beginning of cultivation extended thick and long neurites, compared to those stimulated by DRG-CM (Fig. 5). The combination of the ROCK inhibitor and DRG-CM promoted neuronal differentiation of ES cells when the ROCK inhibitor was added to the culture medium at day 3, but it did not promote neuronal differentiation when added after day 6. The activation of the MAPK signaling pathway is involved in cell survival, differentiation, and growth during neural development (Creedon et al. 1996). Li et al. (2006) reported that the neural differentiation of ES cell-derived neurons was mediated by extracellular signal-regulated kinase (ERK) 1/2 in MAPK family. We reported that the ROCK inhibitor may promote the neuronal differentiation of the ES cells by activating the ERK signaling pathway (Kamishibahara et al. 2014). We found that DRG-CM promoted neurite outgrowth of DRG neurons and contained a small amount of NGF (data not shown). In addition, Ng et al. (2007) reported that DRG-CM contained BDNF. We purported that both DRG-CM containing neurotrophic factors and the ROCK inhibitor may stimulate the ERK signaling pathway. However, the combination of DRG-CM and the ROCK inhibitor did not demonstrate synergistic effects. The ES cells stimulated by DRG-CM at the beginning of cultivation may not be influenced by the ROCK inhibitor. These results showed that the ROCK inhibitor required to be added to the culture medium at the beginning of cultivation for promoting of neuronal differentiation of ES cells.
References
Chang T-C, Chen Y-C, Yang M-H, Chen C-H, Hsing E-W, Ko B-S, Liou J-Y, Wu KK (2011) Rho kinases regulate the renewal and neural differentiation of embryonic stem cells in a cell plating density-dependent manner. PLoS One 5:e9187. doi:10.1371/journal.pone.0009187
Creedon DJ, Johnson EM Jr, Lawrence JC Jr (1996) Mitogen-activated protein kinase-independent pathways mediate the effects of nerve growth factor and cAMP on neuronal survival. J Biol Chem 271:20713–20718
Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154–156. doi:10.1038/292154a0
Gu H, Yu S-P, Gutekunst C-A, Gross RE, Wei L (2013) Inhibition of the Rho signaling pathway improves neurite outgrowth and neuronal differentiation of mouse neural stem cells. Int J Physiol Pathophysiol Pharmacol 5:11–20
Kamishibahara Y, Kawaguchi H, Shimizu N (2014) Promotion of mouse embryonic stem cell differentiation by Rho kinase inhibitor Y-27632. Neurosci Lett 579:58–63. doi:10.1016/j.neulet.2014.07.011
Kitazawa A, Shimizu N (2005) Differentiation of mouse embryonic stem cells into neurons using conditioned medium of dorsal root ganglia. J Biosci Bioeng 100:94–99. doi:10.1263/jbb.100.94
Kitazawa A, Shimizu N (2007) Characterization of neurons differentiated from mouse embryonic stem cells using conditioned medium of dorsal root ganglia. J Biosci Bioeng 104:257–262. doi:10.1263/jbb.104.257
Kitazawa A, Shimizu N (2011) Differentiation of mouse induced pluripotent stem cells into neurons using conditioned medium of dorsal root ganglia. New Biotechnol 28:326–333. doi:10.1016/j.nbt.2011.03.011
Li Z, Theus MH, Wei L (2006) Role of ERK 1/2 signaling in neuronal differentiation of cultured embryonic stem cells. Dev Growth Differ 48:513–523. doi:10.1111/j.1440-169x.2006.00889.x
Martin GR (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 78:7634–7638
Minase T, Ishima T, Itoh K, Hashimoto K (2010) Potentiation of nerve growth factor-induced neurite outgrowth by the ROCK inhibitor Y-27632: a possible role of IP3 receptors. Eur J Pharmacol 648:67–73. doi:10.1016/j.ejphar.2010.09.007
Morizane R, Monkawa T, Itoh H (2009) Differentiation of murine embryonic stem and induced pluripotent stem cells to renal linage in vitro. Biochem Biophys Res Commun 390:1334–1339. doi:10.1016/j.bbrc.2009.10.148
Naka Y, Eda A, Takei H, Shimizu N (2002) Neurite outgrowths of neurons on patterned self-assembled monolayers. J Biosci Bioeng 94:434–439
Nakayama T, Momoki-Soga T, Inoue N (2003) Astrocyte-derived factors instruct differentiation of embryonic stem cells into neurons. Neurosci Res 46:241–249. doi:10.1016/S0168-0102(03)00063-4
Ng BK, Chen L, Mandemakers W, Cosgaya JM, Chan JR (2007) Anterograde transport and secretion of brain-derived neurotrophic factor along sensory axons promote Schwann cell myelination. J Neurosci 27:7597–7603. doi:10.1523/JNEUROSCI.0563-07.2007
Noaksson K, Zoric N, Zeng X, Rao MS, Hyllner J, Semb H, Kubista M, Sartipy P (2005) Monitoring differentiation of human embryonic stem cells using real-time PCR. Stem Cells 23:1460–1467. doi:10.1634/stemcells.2005-0093
Olson MF (2008) Applications for ROCK kinase inhibition. Curr Opin Cell Biol 20:242–248
Pacary E, Legros H, Valable S, Duchatelle P, Lecocq M, Petit E, Nicole O, Bernaudin M (2006) Synergistic effects of CoCl2 and ROCK inhibition on mesenchymal stem cell differentiation into neuron-like cells. J Cell Sci 119:2667–2678. doi:10.1242/jcs.03004
Schmandke A, Schmandke A, Strittmatter SM (2007) ROCK and Rho: biochemistry and neuronal functions of Rho-associated protein kinases. Neuroscientist 13:454–469. doi:10.1177/1073858407303611
Shi J, Wei L (2007) Rho kinase in the regulation of cell death and survival. Arch Immunol Ther Exp 55:61–75. doi:10.1007/s00005-007-0009-7
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676. doi:10.1016/j.cell.2006.07.024
Wobus AM, Grosse R, Schöneich J (1988) Specific effects of nerve growth factor on the differentiation pattern of mouse embryonic stem cells in vitro. Biomed Biochim Acta 47:965–973
Xie C-Q, Huang H, Wei S, Song L-S, Zhang J, Ritchie RP, Chen L, Zhang M, Chen YE (2009) A comparison of murine smooth muscle cells generated from embryonic versus induced pluripotent stem cells. Stem Cells Dev 18:741–748. doi:10.1089/scd.2008.0179
Xue Z-W, Shang X-M, Xu H, Lü S-H, Dong T-W, Liang C-H, Yuan Y (2012) Rho-associated coiled kinase inhibitor Y-27632 promotes neuronal-like differentiation of adult human adipose tissue-derived stem cells. Chin Med J 125:3332–3335. doi:10.3760/cma.j.issn.0366-6999.2012.18.024
Acknowledgments
This work was supported in part by a Grant for the Program for the Strategic Research Foundation at Private Universities S1101017 since April 2011 to one of the authors (N.S.), and by a grant from the Inoue Enryo Memorial Foundation for Promoting Sciences to two of the authors (M. N. and Y. K.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nakamura, M., Kamishibahara, Y., Kitazawa, A. et al. Differentiation patterns of mouse embryonic stem cells and induced pluripotent stem cells into neurons. Cytotechnology 68, 409–417 (2016). https://doi.org/10.1007/s10616-014-9792-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10616-014-9792-2