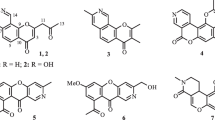
Two new isocoumarins, 5-(2-hydroxyethyl)-7-methoxy-3-(2-oxopropyl)-1H-isocoumarin (1), 7-methoxy-5-methyl-3-(2-oxopropyl)-1H-isocoumarin (2), along with four known ones, were isolated from the fermentation products of an endophytic fungus Aspergillus oryzae derived from cigar tobacco. Their structures were elucidated by spectroscopic methods. Compounds 1 and 2 exhibited anti-TMV activities with inhibition rates of 36.2 and 32.8% at a concentration of 20 μmol/mL, respectively, compared with 35.6% for positive control ningnanmycin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Secondary metabolites are types of natural products that are crucial for growth and development of the source organism. Isocoumarins, isomers of coumarins with an inverted lactone ring, are an important class of naturally occurring metabolites, which are abundantly distributed in natural sources and are being extracted from different plants, molds, lichens, fungi, and bacteria strains [1]. They are abundant in fungal metabolites and mainly derived from the polyketide pathways [2, 3], and the majority of them from fungal sources are always characterized by a carbon substituent at C-3. Isocoumarins also attract much attention from chemists and biologists owing to their diverse structures and biological properties [4, 5].
The genus Aspergillus fungi has attracted considerable attention in recent decades, owing to its ability to metabolize structurally diverse and biologically active products [6, 7]. In previous works, some bioactive metabolites, such as diterpenoids [8], alkaloids [9, 10], butyrolactones [11,12,13], isocoumarins [14,15,16], and the like, have been isolated from the genus of this fungus. With the aim of continuously exploring bioactive metabolites from the genus Aspergillus, the chemical investigations were carried out on the culture broth of the endophytic fungi Aspergillus oryzae obtained from cigar tobacco. As a result, we discovered two undescribed (1 and 2) and four known (3–6) isocoumarins. In addition, compounds 1 and 2 were evaluated for their anti-TMV activities.
The whole culture broth of A. oryzae was extracted with ethyl acetate. The extract was subjected repeatedly to column chromatography on silica gel, MCI, RP-18, and preparative HPLC to afford compounds 1–6, including two new isocoumarins, 5-(2-hydroxyethyl)-7-methoxy-3-(2-oxopropyl)-1H-isocoumarin (1), 7-methoxy-5-methyl-3-(2-oxopropyl)-1H-isocoumarin (2), and four known isocoumarins (3–6). The known compounds, exserolide D (3) [17], oryzaein A (4) [16], tabaisocoumarin A (5) [18], and diaportin (6) [19] were identified by comparison of their spectroscopic data with the literature.
Compound 1 was obtained as a pale-yellow gum. Its molecular formula C15H16O5 was determined by its positive HR-ESI-MS showing a peak at m/z 299.0895 [M + Na]+ (calcd for C15H16NaO5, 299.0889), indicating eight degrees of unsaturation. The IR spectrum showed the absorption bands for carbonyl (1726, 1658 cm–1) and aromatic group (1612, 1568, and 1452 cm–1). The UV spectrum exhibited absorption bands at 210, 267, and 328 nm, also suggesting the existence of an aromatic chromophore. The 1H and 13C NMR spectrum of 1 (Table 1) showed the presence of a 1,2,3,5-tetrasubstituted benzene ring (C-5–C-8, C-4a, C-8a, H-6, and H-8), an 2-oxopropyl group (C-1′–C-3′, H2-1′, and H3-3′) [20], a hydroxyethyl group (C-4′ and C-5′, H2-4′ and H2-5′) [21], a pair of double bonds (C-3, C-4, and H-4), an ester carbonyl carbon (C-1), and a methoxy group (δC 56.0 q and δH 3.77 s). According to the above NMR data (Table 1), the double bonds and ester carbonyl carbon should be incorporated into a benzene ring to form an isocoumarin ring to support the eight degrees of unsaturation, and the above signals can also be attributed to an isocoumarin closely related to oryzaein A [16]. In addition, the existence of the isocoumarin core was also supported by the HMBC correlations (Table 1) from H-4 to C-5, C-4a, C-8a, and from H-8 to C-1.

As the isocoumarin skeleton was determined, the positions of substituents (2-oxopropyl, hydroxyethyl, and methoxy groups) can also be determined by further analysis of its HMBC data (Table 1). The HMBC correlations from the H2-1′ to C-3, C-4, from H-4 to C-1′ established that the 2-oxopropyl group was located at C-3. The methoxy group located at C-7 was clearly indicated by the HMBC correlations from methoxy proton (δH 3.77 s) to C-7. Furthermore, the hydroxyethyl group located at C-5 can also be determined by the HMBC correlations of H2-4′ with C-5, C-6, C-4a, and of H-5′ with C-5. On the basis of the above evidence, the structure of 1 was established as shown, and gave the systematic name of 5-(2-hydroxyethyl)-7-methoxy-3-(2-oxopropyl)-1H-isocoumarin.
Compound 2 was also obtained as a pale-yellow gum. The molecular formula was established as C14H14O4, by the [M + Na]+ ion at m/z 269.0796 (calcd for C14H14NaO4, 269.0790) in HR-ESI-MS. The 1H and 13C NMR data (Table 1) of 1 and 2 showed high similarity in C-1–C-8, C-1′–C-3′, C-4a, C-8a, and OMe, with the only obvious difference of the extra presence of a methyl group (C-4′ and H3-4′) and the disappearance of a hydroxyethyl group, which suggested that 2 was an isocoumarin analog. The methyl group located at C-5 in the isocoumarin skeleton was determined by the HMBC correlations from H3-4′ to C-5, C-6, C-4a, and from H-6 to C-4′. In addition, the positions of the 2-oxopropyl and methoxy groups can also be determined by further analysis of their HMBC correlations. Thus, the structure of 2 was determined to be 7-methoxy-5-methyl-3-(2-oxopropyl)-1H-isocoumarin.
As the isocoumarin derivatives from fungus exhibit potential anti-virus [14, 15, 22], compounds 1 and 2 were also evaluated for their anti-TMV activities. The anti-TMV activity was tested by the half-leaf method [23, 24], using ningnanmycin as a positive control. The results showed that compounds 1 and 2 exhibited potential anti-TMV activity, with inhibition rates of 36.2 and 32.8% respectively, and the positive control showed an inhibition rate of 35.6%.
EXPERIMENTAL
General Methods. UV spectra were obtained using a Shimadzu UV-1900 spectrophotometer. A Bio-Rad FTS185 spectrophotometer was used for scanning IR spectra. 1H, 13C, and 2D NMR spectroscopic data were recorded on a DRX-500 NMR spectrometer with TMS as the internal standard. ESI-MS and HR-ESI-MS analyses were measured on an Agilent 1290UPLC/6540 Q-TOF mass spectrometer. Semipreparative HPLC was performed on an Agilent 1260 preparative liquid chromatograph with Zorbax PrepHT GF (2.12 mm × 25 cm) or Venusil MP C18 (2.0 mm × 25 cm) columns. Column chromatography was performed using silica gel (200–300 mesh, Qingdao Marine Chemical, Inc., Qingdao, China), Lichroprep RP-18 gel (40–63 μm, Merck, Darmstadt, Germany), Sephadex LH-20 (Sigma-Aldrich, Inc., USA), or MCI gel (75–150 μm, Mitsubishi Chemical Corporation, Tokyo, Japan).
Fungal Material. The culture of Aspergillus oryzae (YNCA-20-32) was isolated from the leaves of cigar tobacco, collected from Gengma County, Lincang Prefecture, Yunnan Province, in 2020. The strain was identified by one of authors (Dr. Yu-Ping Wu) based on the analysis of its sequence. It was cultivated at room temperature for 7 days on potato dextrose agar at 28°C. Agar plugs were inoculated into 250-mL Erlenmeyer flasks, each containing 100 mL potato dextrose broth and cultured at 28°C on a rotary shaker at 180 rpm for 5 days. Large-scale fermentation was carried out in 100 Fernbach flasks (500 mL) each containing 300 mL of medium (glucose 5%; peptone 0.15%; yeast 0.5%; KH2PO4 0.05%; MgSO4 0.05% in 1.0 L of deionized water; pH 6.5 before autoclaving). Each flask was inoculated with 5.0 mL of cultured broth and incubated at 27°C for 20 days.
Extraction and Isolation. The whole culture broth of A. oryzae was extracted four times with ethyl acetate (4 × 10 L) at room temperature and filtered. The crude extract (136 g) was applied to silica gel column chromatography, eluting with a CHCl3–(CH3)2CO gradient system (9:1, 8:2, 7:3, 6:4, 5:5). Five fractions were obtained from the silica gel column and individually decolorized on MCI gel to yield fractions A–E. The further separation of Fr. B (8:2, 12.5 g) by silica gel column chromatography, eluted with petroleum ether–EtOAc (9:1, 8:2, 7:3, 6:4, 1:1), yielded subfractions B1–B5. Subfraction B3 (7:3, 3.22 g) was subjected to RP-18 column chromatography (MeOH–H2O, 20:80–80:20 gradient) and HPLC to give 4 (18.6 mg). The further separation of Fr. C (7:3, 9.85 g) by silica gel column chromatography, eluted with petroleum ether–EtOAc (8:2, 7:3, 6:4, 1:1), yielded subfraction C1–C4. Subfraction C3 (6:4, 2.87 g) was subjected to RP-18 column chromatography and HPLC (MeOH–H2O, 20:80–60:40 gradient) to give 1 (18.5 mg), 2 (16.4 mg), 3 (12.8 mg), and 5 (20.6 mg). The further separation of fraction D (6:4, 12.2 g) by silica gel column chromatography, eluted with petroleum ether–EtOAc (7:3, 6:4, 1:1, 2:1), yielded subfraction D1–D4. Subfraction D3 (1:1, 2.24 g) was subjected to RP-18 column chromatography and HPLC (MeOH–H2O, 20:80–60:40 gradient) to give 6 (14.5 mg).
5-(2-Hydroxyethyl)-7-methoxy-3-(2-oxopropyl)-1H-isocoumarin (1), obtained as a pale yellow gum, C15H16O5. UV (MeOH, λmax, nm) (log ε): 210 (3.90), 267 (3.59), and 328 (3.44). IR (KBr, νmax, cm–1): 3416, 3075, 2952, 2857, 1726, 1658, 1612, 1568, 1452, 1369, 1142, 1070. For 1H (500 MHz) and 13C (125 MHz) NMR spectra (CDCl3), see Table 1. ESI-MS m/z 299 [M + Na]+; HR-ESI-MS m/z 299.0895 [M + Na]+ (calcd for C15H16NaO5, 299.0889).
7-Methoxy-5-methyl-3-(2-oxopropyl)-1H-isocoumarin (2), obtained as a pale yellow gum, C14H14O4. UV (MeOH, λmax, nm) (log ε): 210 (3.92), 265 (3.61), 326 (3.42). IR (KBr, νmax, cm–1): 3082, 2940, 2861, 1725, 1662, 1615, 1564, 1443, 1362, 1138, 1064. For 1H and 13C NMR (500 and 125 MHz, CDCl3), see Table 1. ESI-MS m/z 269 [M + Na]+; HR-ESI-MS m/z 269.0796 [M + Na]+ (calcd for C14H14NaO4, 269.0790).
Anti-TMV Assays. The anti-TMV activities were tested using the half-leaf method [23, 24], and ningnanmycin (2% water solution), a commercial product for plant disease in China, was used as a positive control. The virus was inhibited by mixing with a solution of the tested compounds. After 30 min, the mixture was inoculated on the left side of the stems of Nicotiana glutinosa, whereas the right side of the leaves was inoculated with the mixture of DMSO solution and the virus as a control. The local lesion numbers were recorded 3–4 days after inoculation. Three repetitions were conducted for each compound. The inhibition rates were calculated according to the formula:
where C is the average number of local lesions of the control and T is the average number of local lesions of the treatment.
References
G. Shabir, A. Saeed, and H. R. El-Seedi, Phytochemistry, 181, 112568 (2021).
A. Saeed, Eur. J. Med. Chem., 116, 290 (2016).
D. Engelmeier, F. Hadacek, O. Hofer, G. Lutz-Kutschera, M. Nagl, G. Wurz, and H Greger, J. Nat. Prod., 67, 19 (2004).
A. Saeed, Eur. J. Med. Chem., 116, 290 (2016).
S. Pal, V. Chatare, and M. Pal, Curr. Org. Chem., 15, 782 (2011).
S. Skanda and B. S. Vijayakumar, Curr. Microbiol., 78, 1317 (2021).
M. Y. Deng, X. Chen, Z. Y. Shi, and S. S. Xie, Fitoterapia, 151, 104882 (2021).
W. G. Wang, L. Q. Du, S. L. Sheng, A. Li, Y. P. Li, G. G. Cheng, G. P. Li, G. L. Sun, Q. F. Hu, and Y. Matsuda, Org. Chem. Front., 6, 571 (2019).
M. Zhou, M. M. Miao, G. Du, X. N. Li, S. Z. Shang, W. Zhao, Z. H. Liu, G. Y. Yang, C. T. Che, Q. F. Hu, and X. M. Gao, Org. Lett., 16, 5016 (2014).
G. Y. Yang, J. M. Dai, Q. L. Mi, Z. J. Li, Xue-Mei Li, J. D. Zhang, J. Wang, Y. K. Li, W. G. Wang, M. Zhou, and Q. F. Hu, Phytochemistry, 198, 113137 (2022).
K Zhou, L. Zhu, X. L. Wang, T. D. Zhang, Y. D. Wang, W. Dong, B. K. Ji, H. Y. Yang, G. Du, Q. F. Hu, and M. Zhou, Chem. Nat. Compd., 52, 591 (2016).
L. Yuan, W. Z. Huang, K. Zhou, Y. D. Wang, W. Dong, G. Du, X. M. Gao, Y. H. Ma, and Q. F. Hu, Nat. Prod. Res., 29, 1914 (2015).
M. Zhou, G. Du, H. Y. Yang, C. F. Xia, J. X. Yang, Y. Q. Ye, X. M. Gao, X. N. Li, and Q. F. Hu, Planta Med., 81, 235 (2015).
G. Du, Z. C. Wang, Y. K. Yang, H. M. Yang, H. Y. Yang, M. Zhou, Y. Q. Ye, X. M. Li, and Q. F. Hu, Heterocycles, 91, 1996 (2015).
M. Zhou, K. Zhou, P. He, K. M. Wang, R. Z. Zhu, Y. D. Wang, W. Dong, G. P. Li, H. Y. Yang, Y. Q. Ye, G. Du, X. M. Li, and Q. F. Hu, Planta Med., 82, 414 (2016).
M. Zhou, J. Lou, Y. K. Li, Y. D. Wang, K. Zhou, B. K. Ji, W. Dong, X. M. Gao, G. Du, and Q. F. Hu, Arch. Pharm. Res., 40, 32 (2017).
R. X. Li, S. X. Chen, S. B. Niu, L. D. Guo, J. Yin, and Y. S. Che, Fitoterapia, 96, 88 (2014).
S. Z. Shang, W. X. Xu, L. Li, J. G. Tang, W. Zhao, P. Lei, M. M. Miao, H. D. Sun, J. X. Pu, Y. K. Chen, and G. Y. Yang, Phytochem. Lett., 11, 53 (2015).
P. G. Mantle, Phytochemistry, 57, 165 (2001).
Q. L. Mi, W. S. Kong, Y. K. Li, X. Liu, W. L. Zeng, H. Y. Xiang, M. Dong, G. Y. Yang, Q. F. Hu, W. G. Wang, Q. Gao, and M. Zhou, Chem. Nat. Compd., 57, 432 (2021).
D. W. Wang, F. Wu, Y. N. Zhu, L. Liu, D. Miao, M. Zhou, W. G. Wang, Q. F. Hu, and J. B. Yue, Chem. Nat. Compd., 57, 462 (2021).
Q. F. Hu, H. H. Xing, Y. D. Wang, Z. H. Yu, K. L. Yan, K. Zhou, W. Dong, M. Zhou, H. Y. Yang, D. L. Zhu, and G. Du, Chem. Nat. Compd., 53, 436 (2017).
Q. F. Hu, B. Zhou, J. M. Huang, X. M. Gao, L. D. Shu, G. Y. Yang, and C. T. Che, J. Nat. Prod., 76, 292 (2013).
M. Zhou, M. M. Miao, G. Du, S. Z. Shang, W. Zhao, Z. H. Liu, G. Y. Yang, C. T. Che, Q. F. Hu, and X. M. Gao, Org. Lett., 16, 5016 (2014).
Acknowledgment
This project was supported financially by China Tobacco Monopoly Bureau Grants and Yunnan Provincial Tobacco Monopoly Bureau Grants (110202103018, 2022530000241002), the National Natural Science Foundation of China (No. 21967021), and the Foundation of Yunnan Innovative Research Team (2019HC020).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Published in Khimiya Prirodnykh Soedinenii, No. 2, March–April, 2023, pp. 206–209.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kong, GH., Gu, XJ., Wu, J. et al. Antiviral Isocoumarins from a Cigar Tobacco-Derived Endophytic Fungus Aspergillus oryzae. Chem Nat Compd 59, 242–245 (2023). https://doi.org/10.1007/s10600-023-03966-0
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10600-023-03966-0