Abstract
Heart disease is a complex disease, and many people around the world suffer from this disease. Due to the lack of a healthy lifestyle, it is the most common cause of death worldwide. Machine learning plays an important role in medical treatment. The goal of this research is to develop a machine learning model to help diagnose heart disease quickly and accurately. In this article, an effective and improved machine learning method is proposed to diagnose heart disease. We designed a novel and robust ensemble model that combines the top three classifiers, namely Random Forest, XGBoost and Gradient Boosting Machine, to effectively diagnose heart disease. We used an ensemble voting method to combine the results of the top three classifiers to improve the prediction of heart disease. We used a combined heart disease dataset containing five different datasets (Hungary, Statlog, Switzerland, VA Long Beach and Cleveland). Feature selection algorithms (Pearson Correlation, Univariate Feature Selection, Recursive Feature Elimination, Boruta Feature Selection, Random forest, and LightGBM) are used to select highly relevant features based on rankings to improve classification accuracy. The proposed ensemble model is designed using seven highly relevant features, and a comparison of machine learning algorithms and ensemble learning techniques is applied to the selected features. Different performance evaluation methods are used to evaluate the proposed model: accuracy, sensitivity, precision, F1-score, MCC, NPV and AUC. Results analysis shows that the ensemble model achieves excellent classification accuracy, sensitivity, and precision of 96.17%, 98.37%, and 94.53%. Our proposed model performs better than existing models and individual classifiers. The results show that the proposed ensemble method can effectively predict the risk of heart disease.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
In today’s world, heart disease is complex and one of the leading causes of death in the world. According to the World Health Organization (WHO) [1], 17.7 million people died in 2016 from heart disease, which accounts for about 31% of the world’s deaths. A huge number of deaths are common in low- and middle-income countries. In the United States and other developed countries, about half of all deaths are caused by heart disease. Also, one-third of the deaths of all people in the world are associated with heart disease. Heart disease affects the regular function of the heart. Blockage of the coronary arteries is the most common cause of heart disease because it supplies blood to the heart itself. It is estimated that over 30 million people will die from heart disease by 2040. Heart diseases affect the country’s economy as well as human health. The increasing incidence of high-mortality cardiovascular disease poses significant risks and burdens to healthcare systems around the world. Researchers are trying to find an effective method for detecting heart disease because current methods of diagnosing heart disease are not very effective at early detection for several reasons, such as accuracy [2]. Several tests are required to predict heart disease. Lack of experience of medical personnel can lead to erroneous predictions [3]. Diagnosis and treatment of heart disease can be very difficult, especially in developing countries where the trained medical professionals and other resources needed for proper diagnosis and care of patients with heart disease are lacking [4]. In this case, early diagnosis of heart disease and proper dosing will improve patient safety.
Machine learning (ML) techniques are applied in many fields of medical science. However, researchers are always looking for ways to optimize these techniques. Ensemble learning is one such approach that has been shown to improve ML problems [5]. An ensemble classifier is a process of combining multiple models (such as majority voting) to solve specific computational intelligence problems. Research shows that ensemble classifiers generally have better performance than conventional classifiers [6]. ML prediction models require appropriate data for training and testing. When trained with appropriate data, ML algorithms can effectively identify diseases [7]. The performance of a ML model can be improved by using a balanced dataset to train and test the model. In addition, the predictive capabilities of the model can be improved by using the appropriate relevant features from the data. Therefore, feature selection and data balancing are very important to improve model performance. It is essential to select important features that can be used as risk factors in the prediction. The right combination of features helps to develop accurate predictive models. Researchers have proposed various diagnostic methods, but these methods do not effectively diagnose heart disease. Data preprocessing is a crucial step, and it is essential for data standardization to improve predictability.
In order to obtain good results, a suitable ML model must be used. Obviously, a good L model is one that works well not only with training data, but also with testing data. In addition, when the model is trained and tested on the data set, appropriate feature selection techniques and performance evaluation metrics are critical to the model. ML techniques can be used to identify several diseases, but the diagnosis of heart disease is the main goal of this article. Researchers have applied a variety of techniques to predict heart disease, either as individual classifiers or hybrid [8]. There are still many problems that may not accurately predict heart disease, such as limited medical data sets, feature selection and ML techniques. One of the major issues encountered in previous approaches is low accuracy, which may be attributed to the utilization of irrelevant features in the dataset. To address these problems and improve the accuracy of heart disease detection, new methods are required.
In our previous study in May 2021 [9], we used six ML algorithms and several evaluation techniques were used to evaluate the performance of each algorithm. We used UCI Cleveland heart disease dataset for the training and testing purpose. We proposed ensemble classifier by combining KNN, XGBoost and Logistic Regression and yields the accuracy of 92%, precision of 91.1%, recall of 94% and F1-score of 93%. Since the data in previous work was limited, this motivated us to further research and develop a more robust model with reduced feature set based on the ensemble learning technique by increasing the amount of data to identify patients infected with heart disease to help healthcare professionals. This work is an updated version of our previous work, but the method has been greatly expanded and improved. Ensemble Algorithm and several algorithms, such as logistic regression (LR), XG Boost (XGB), k nearest neighbor (KNN), gradient boosting machine (GBM), decision tree (DT), AdaBoost, Naive Bayes (NB) and Random Forest (RF) are used to classify whether people have heart disease. The purpose of this study is to develop effective methods for predicting heart disease as accurately as possible. The main contributions of this research are summarized as follows:
-
Five publicly available datasets are combined to create a larger, more reliable dataset that is used for evaluation, and comparisons are made using existing state of art methods.
-
Nine different state-of-the-art ML methods are tested for heart disease classification: k-nearest neighbors (KNN), XG boost (XGB), logistic regression (LR), gradient boosting machine (GBM), Multi-Layer Perceptron (MLP), decision Tree (DT), Naive Bayes (NB), AdaBoost and Random Forest (RF).
-
The heart disease diagnostic dataset has 11 features. Feature selection techniques are used to select important features from a dataset for automated heart disease diagnosis.
-
For comparison, all ML classifiers are evaluated for comparative studies using evaluation metrics such as accuracy, sensitivity, precision, F1-score, Matthews’s correlation coefficient (MCC), negative predictive value (NPV), and area under the curve.
-
We have proposed ensemble classifier with a reduced subset of features constructed using three classifiers, namely Random Forest, XGBoost, and GBM, and the results of the classifiers are combined using a voting method to predict early stage heart disease.
-
A comparative study is conducted with recent scholarly works and other well-known ML algorithms.
2 Related work
Artificial intelligence and ML algorithm applications have become very popular in recent years due to their improved performance in making predictions. ML models are widely used to predict and identify the risk of heart disease. Smoking, age, tobacco, high blood cholesterol, diabetes, overweight, and high blood pressure are considered risk factors for heart and vascular disease [10]. To accurately and efficiently predict coronary heart disease, researchers consider a variety of ML-based diagnostic techniques for predicting heart disease, either as individual classifiers or ensemble classifiers. This study presents some of the existing L diagnostic methods carried out on the classification of heart disease to explain the importance of the proposed work.
Amin et al. [11] used a variety of feature extraction methods to identify key features for predicting heart disease. Atallah and Al-Mousa [12] proposed ensemble learning model based on different ML classifiers to predict heart disease. The best accuracy achieved is 90%. Kannan and Vasanthi [13] predicted heart disease using four ML algorithms: and achieved the highest classification accuracy of 86.5%. Gudadhe et al. [14] used multilayer Perceptron and SVM algorithms for a heart disease diagnostic system and achieved an accuracy of 80.41%. Prasad et al. [15] proposed a LR predictive mechanism for heart disease and achieved an accuracy of 79%, while KNN achieved an accuracy of 78%. Melillo et al. [16] proposed an automatic classifier to classify patients with heart disease. In their study, CART performed best with a sensitivity of 93.3% and a specificity of 63.5%. Nalluri et al. [17] used two ML classifiers to improve the classification accuracy. The results show that the accuracy of LR is 85.68%, which is better than XGBoost, reaching an accuracy of 84.46%. Sapra et al. [18] uses two data sets (Z-Alizadesh Sani and Cleveland Heart Disease Data Set), which are used to train ML classifiers for the prediction of heart disease. The results show that compared with other algorithms, the gradient boosting tree achieves the best accuracy of 84%. Raza proposed an ensemble architecture using majority voting [19]. It combines LR, MLP and NB to predict the patient’s heart disease. The ensemble method achieves 88.88% classification accuracy. Mohan et al. [20] combined RF with linear models and proposed an ensemble method. The proposed method achieves a classification accuracy of 88.7% on the test set. The author in [21] proposed a hybrid PSO algorithm for diagnosing heart disease. This study used 13 feature attributes to predict heart disease with an accuracy of 84.25%. Geweid and Abdallah [22] used ECG signals to design an optimized and improved SVM model for the identification of heart diseases. Haq et al. [23] used seven ML algorithms with three feature selection methods for predicting heart disease. The highest classification accuracy of 89% was achieved. Rashmi et al. [24] experimented with the Cleveland dataset. The proposed DT algorithm obtained an accuracy of 75.55%. Sharma et al. [25] and Dwivedi et al. [26] used the same data set to train the KNN algorithm. They have achieved classification accuracy of 90.16% and 80% respectively. The author of [27] used SMO, NN, bagging and NB. SMO achieved the highest accuracy of 94.08%. Guidi et al. [28] used different ML classifiers in their research and compared their performance. Random forest and CART have the best results, with an accuracy of 87.6%. Liu et al. [10] have proposed a classification system for heart disease using the rough set technique. The proposed method provided a classification accuracy of 92.32%. The author in [29] proposed a nested ensemble algorithm with feature selection and data balance. The classification accuracy of the algorithm on the balanced data set reached 94.66%. In [30], multiple feature selection algorithm are used to propose an ensemble model, which achieved 93.7% accuracy in the prediction of heart disease. Abdar et al. [31] proposed a novel method called N2Genetic optimizer. Then, nuSVM is used to predict heart disease patients. The accuracy of the proposed detection method on the data set is 93.08%. Shah et al. [32] proposed a system to study different conditions that may affect the heart and the main factors of death. DT, NB, RF and KNN are used. Of the 76 attributes, only 14 attributes were used. KNN performs better than other classifiers. Latha and Jeeva [33] developed an ensemble architecture to improve the accuracy of heart disease prediction. Their approach yielded a significant improvement, achieving an accuracy of 85.48%. E. Nasarian et al. [34] introduced a hybrid feature selection algorithm for coronary artery disease. To address the issue of data imbalance, they applied an oversampling method. Their approach resulted in an impressive accuracy of 92.58%.
All studies presented in the review used different methods for detecting heart disease in the early stages. However, all of these methods lack the accuracy of predicting heart disease. The accuracy of the heart disease classifier needs to be further improved for accurate detection at an early stage. Due to the use of irrelevant features in the data set, the previous methods are less accurate. To address these limitations, the aim of this study is to develop an ensemble classifier with a reduced feature dataset built using three base classifiers, namely XGBoost, Random forest and Gradient Boosting Machine (GBM) for predicting heart disease. Experiments are also performed to identify the most important features using a variety of feature selection techniques. Publicly available heart disease datasets are used for evaluation and compare the performance achieved by the proposed classifier with existing state-of-the-art methods.
3 Materials and methods
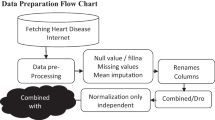
The purpose of this study is to improve the classification performance of patients suffering from heart disease. Figure 1 illustrates the workflow of the proposed ensemble classification algorithm for the prediction of heart disease. It consists of various submodules including data collection, data preprocessing, model training and testing, feature selection, ensemble model prediction and performance evaluation. The five heart disease data sets discussed later are integrated into one training data set. First, we apply nine modern classification algorithms separately. The feature selection methods are then used separately to find the most important features. After feature selection, ML classifiers are trained to evaluate the performance of the classifiers using the selected features. Three classification algorithms are selected based on the best performance to propose a new ensemble classification method. The classification results of the three best classifiers (XGBoost, RF and GBM) are combined with a ensemble technique for the final classification result of heart disease.
3.1 Dataset
The heart disease data set used in this study was collected from the ML database [35] of the University of California, Irvine (UCI). The heart disease dataset [36] is used to train and evaluate the models. This heart disease dataset is curated by combining five popular heart disease datasets that are already available independently (Cleveland, Hungarian, Switzerland, VA Long Beach and Statlog). The dataset contains medical records of 1190 patients, and all patient profiles have 11 clinical features and one target variable. Table 1 depicts the features and complete description of the data set.
3.2 Data preprocessing
The performance of any classification problem depends on the standard of the data set. The real-time dataset requires preprocessing before training models to make predictions robust. Preprocessing techniques (such as data splitting, data standardization, handling missing values) have been applied to the data set.
3.2.1 Handling missing values
During data preprocessing, missing values in the combined dataset are analyzed. We found that the heart disease dataset contained some missing values across individual features. There are many ways to deal with missing values, such as ignoring the missing value completely and replacing the missing value with some numeric value. These data need to be cleaned up for noise and missing values for accurate and effective results. Therefore, we deleted 18 records with missing values.
3.2.2 Data standardization
Data sets need to be standardized when the units of measure are different. By applying the min-max scaling technique, the entire data set can be normalized within the minimum and maximum scaling range of each feature. The main goal is that all features are in the range of 0 to 1.
3.2.3 Data splitting
In this step, 80% (937 instances) of the heart disease dataset is used to train the models, and the remaining 20% (235 instances) is used to evaluate the performance of the models on the heart disease data set.
3.3 Feature selection techniques
Feature selection is very important in handling redundant features. This is the concept of improving model performance through the use of ML. Some irrelevant input features can reduce the performance of the classifier. Therefore, it is difficult to select important features for the prediction task from a particular set of features. In this paper, we used six feature selection methods (Recursive feature selection, Univariate feature selection, Pearson correlation, Boruta feature selection, Random forest, LightGBM) to select related features from a dataset.
3.3.1 Recursive Feature Elimination (RFE)
RFE is a popular wrapper-based feature selection method that also uses filter-based feature selection internally. RFE is popular because it is effective in selecting those features in the training dataset that are more important in predicting the target variable. The working principle of RFE is to start with all the features in the training data set, and then remove the weakest feature until it has a specified number of features. This is achieved by first building a model on a complete set of elements, ranking the elements by importance, eliminating the weakest elements and rebuilding the model. Repeat this process until the specified number of features are retained.
3.3.2 Univariate feature selection
Univariates or \({x}^{2}\)(chi-square) are filter-based feature selection method and only indicate the importance of each of the original features. We compute the chi-square to select the features between each feature and the target class, and select the number of features with the best chi-square score. Based on this, we can choose to keep the most important features and discard the least important ones. It is very fast in calculation and provides a way to quickly filter features.
3.3.3 Pearson correlation
The Pearson correlation coefficient (PCC) is a filter-based feature selection method used to measure the strength of the linear relationship between two sets of data. It is known as the best way to find the relationship between two variables because it is based on the method of covariance. The PCC value ranges from − 1 to + 1. 1 represents positive correlation, -1 represents negative correlation, and 0 means no correlation.
3.3.4 Boruta feature selection
Boruta is a wrapper algorithm for all relevant feature selection built around the RF algorithm [37]. The RF classification algorithm can usually be executed without setting parameters. First, it duplicates the dataset and creates shuffled copies of all features, which are called shadow features. Then train a classifier, such as a RF classifier, on the dataset and apply feature importance measurements such as mean decrease accuracy to assess the importance of each feature. The Boruta algorithm always removes features that are considered insignificant. On the other hand, features that are far superior to shadows are admitted to be confirmed.
The Boruta algorithm includes the following steps:
-
1.
Add shadow attributes and shuffle the values in each column.
-
2.
During each iteration, the Z-scores of the shuffled copies and original features are evaluated to see if the latter perform better than the former. In this case, the algorithm will mark the feature as important.
-
3.
Features whose importance is much lower than the Z score are considered “unimportant” and are permanently deleted from the information system.
-
4.
The feature becomes important when its Z-score is higher.
-
5.
Remove all shadow attributes.
-
6.
Finally, when all the features are confirmed or rejected, the Boruta algorithm will stop.
3.3.5 Random Forest (RF)
Random forests also provide excellent feature selection indicators. It is the most flexible and easy-to-use algorithm. Random forest is a collection of multiple decision trees. This is a better correlation integration method than deep decision trees and avoids overfitting by generating trees on random subsets. Random forest uses Gini index or mean decrease impurity (MDI) to calculates each feature’s importance. Gini Importance or MDI will calculate the importance of each feature as the sum of the number of samples that contain the feature, and it is proportional to the number of samples it divides. The more the Gini index decreases for a feature, the more significant the feature is.
3.3.6 LightGBM
LightGBM (Light Gradient Boosting Machine) is a fast and effective framework that uses tree-based learning algorithms. Unlike other ensemble techniques, LGBM grows trees leaf by leaf, reducing losses during the sequential boost process. The importance of each feature can be derived from the feature_importances_ attribute built into the algorithm. LightGBM accelerates learning rates and reduce memory usage by segmenting the values of sequential features.
3.4 Machine learning classifiers
This section discusses various ML classifiers used to predict results based on data. This work proposes a technique that uses classification methods to predict heart disease, and uses an ensemble of classifiers to improve the classification accuracy. The details of several ML classifiers are discussed below.
3.4.1 K-Nearest Neighbors (KNN)
KNN is one of the simplest ML algorithms based on supervised learning techniques. It classifies objects based on the nearest neighbor. This is a nonparametric algorithm, which means that it does not assume the underlying data. It is called the Lazy Learner algorithm because it does not learn immediately and can be used for classification and regression. In KNN, k is used to indicate the number of nearest neighbors, and better performance can be obtained by choosing a suitable value of k.
3.4.2 Random Forest (RF)
RF named random decision forests to perform a ML role that can be used to solve classification and regression problems. RF is an easy-to-use algorithm that produces excellent results without adjusting hyperparameters in most cases. It contains many decision trees on each subset of a given data set and averages them to improve performance. It is an effective method for assessing missing data and maintaining accuracy when most of the data is missing.
3.4.3 Decision Tree Classifier (DTC)
Decision trees can be used to solve regression and classification problems. In the DTC, we start with the root node and then we expand it to more branches and hence it forms a tree-like structure. In a decision tree (DTC), if we want to make a prediction of the class from a given dataset. We start from the root node, and then the algorithm makes the comparison of the values of the root attribute with the real dataset attribute. On the basis of this comparison, the algorithm decides the branch and then moves to the next node. From the nested node, it again makes a comparison of the attribute values with the other sub-nodes and then jumps next. This process continues until it reaches the leaf node of the tree.
3.4.4 XGBoost
XGBoost is an implementation of gradient boosting decision tree, which aims to improve efficiency and compactness. XGBoost provides a parallel tree boosting, which can quickly and accurately solve many data science problems. When it comes to computational efficiency (memory usage, speed, performance, etc.), XGBoost is better than GBM. In addition, it utilizes a more normalized model, which minimizes model complexity and improves prediction accuracy. It has a wide range of configurable parameters and is effective with large datasets. XGBoost does not need normalized features. If the data is non-linear, it can work well.
3.4.5 Gradient Boosting Machine (GBM)
Gradient boosting is mostly used for classification problems where many weak classifiers work together to create powerful learning model. GBM consists of three main components: a loss function calculated by gradient descent method, a weak learner for prediction, and an additive model to minimize losses by implementing more decision trees. It can deal with missing data values very efficiently.
3.4.6 Multilayer Perceptron (MLP)
MLP is a deep artificial neural network that contains many perceptron’s. It traditionally consists of three groups of layers: an input layer for receiving a signal, an output layer that makes a decision, but may have several hidden layers in between the aforementioned layers, which are the true computational mechanism of MLP. MLPs are applied to supervised learning problems and often train models based on the correlation between input and output variables. MLP is designed to approximate any continuous function and classify data sets that cannot be linearly separated. The main purpose of MLP is classification, recognition and prediction.
3.4.7 Logistic Regression (LR)
LR is one of the most common and useful classification algorithms in ML and belongs to a supervised learning technique. LR predicts the output of a categorical dependent variable, it works when the dependent variable is 0 or 1, which contains 1 for a positive class and 0 for a negative class. Linear and logistic regressions differ in the dependent variable. Linear regression is a more suitable method for continuous variables. In LR, we fit an S-shaped logistic function that predicts two maximum values. It is an important algorithm because it can use continuous data sets to determine probabilities and classify new data.
3.4.8 AdaBoost
The AdaBoost algorithm or Adaptive Boosting is a boosting technique used to improve the performance of decision trees, which is based on the problem of binary classification. Weak models are trained using weighted training data and added sequentially to make the classifier more robust. One of the main advantages of the AdaBoost algorithm is its speed, simplicity and ease of programming. This is an algorithm for constructing a “strong” classifier in the form of a linear combination of simple weak classifiers.
3.4.9 Naïve Bayes
Naive Bayes classification algorithms are generally known for its simplicity and effectiveness. This is a supervised learning algorithm used to solve classification problems based on Bayes’ theorem. It can be constructed quickly and make quick predictions. It is scalable, requiring multiple linear parameters for different parameters in the training task. It is a probabilistic classifier, which learns the probabilities of the features according to the target class.
3.5 Hyperparameter tuning
In our proposed system, the best training hyperparameters for each classifier are obtained using GridSearchCV to get higher accuracy. The GridSearchCV is used to train a ML model with multiple combinations of training hyperparameters, and the best parameter values are extracted, which optimizes the evaluation metric. Tuning is a fundamental part of all types of classifiers to get accurate predictions. As a result, we configured our 9 classifiers including KNN, RF, DTC, XGB, GBM, MLP, LR, AdaBoost and NB. The model parameters used are shown in Table 2.
3.6 Proposed ensemble method for predicting heart disease
An ensemble is a ML model that combines predictions from two or more models. This is an effective classification method that combines weak classifiers with strong classifiers to improve efficiency. The utilization of ensemble models, as opposed to single models, offers several advantages, including enhanced prediction accuracy and improved overall performance. In this paper, we propose a novel ensemble method specifically designed for diagnosing heart disease. Our approach involves the selection of relevant attributes followed by a comprehensive evaluation of nine ML classification algorithms for heart disease classification. These classifiers are rigorously assessed, and based on their accuracy, we identify the three most promising classification algorithms for inclusion in our ensemble model. To construct our ensemble model, we employ a voting ensemble technique that combines the results of the selected classifiers to determine the final classification outcome for heart disease diagnosis. We investigate two distinct voting mechanisms: majority-based voting and soft voting. Majority-based voting involves aggregating the predictions from the three selected classifiers and selecting the class that receives the majority of votes. On the other hand, soft voting assigns weights to each classifier’s predictions based on their confidence levels, and the final classification decision is made by summing these weighted predictions. To implement the ensemble model, each selected classifier undergoes a comprehensive training process using a suitable dataset for heart disease diagnosis. We fine-tune the hyperparameters of each classifier to optimize their performance. The individual classifiers are then combined within the ensemble framework, leveraging their collective intelligence to produce a more robust and accurate prediction model. The primary objective of combining multiple algorithms within our ensemble framework, as opposed to relying solely on individual classifiers, is to harness the complementary strengths of each algorithm. By exploiting the diversity of the selected classifiers, we aim to mitigate the limitations and biases inherent in any single classifier. The ensemble model capitalizes on the varying decision boundaries and feature representations learned by each classifier, ultimately leading to improved overall performance and diagnostic accuracy for heart disease.
We evaluate the performance of our ensemble model using a comprehensive set of evaluation metrics specifically tailored for heart disease diagnosis. These metrics include but are not limited to accuracy, precision, recall, and F1 score. By considering multiple evaluation metrics, we gain a holistic understanding of the ensemble model’s performance and its capability to accurately predict the presence or absence of heart disease. Overall, our proposed ensemble model represents a significant advancement in the diagnosis of heart disease by effectively combining the strengths of multiple ML classifiers. The comprehensive evaluation, selection, and integration of the classifiers within the ensemble framework, along with the utilization of voting mechanisms, contribute to superior performance and improved accuracy in heart disease diagnosis. Figure 2 shows how the ensemble approach is being used to improve the diagnosis of heart disease.
3.6.1 Majority voting
In majority voting, the classification of samples is determined by aggregating the votes received from multiple classifiers, denoted as C, for a specific class. The category label y is predicted based on the mode of the individual classifier predictions. This can be mathematically expressed using Eq. 1.
By employing the majority voting mechanism, the ensemble model takes advantage of the collective decision-making of the individual classifiers. It leverages the idea that diverse classifiers may capture different aspects of the data and contribute varied insights, leading to a more robust and accurate prediction.
3.6.2 Soft voting
In soft voting, the output class prediction is determined by averaging the probabilities assigned by each individual classifier. The category label y is calculated as the average of the predictions from all classifiers, as shown in Eq. 2. By using soft voting, the ensemble model considers the confidence levels or probabilities of the individual classifier predictions, rather than simply the class labels. This approach considers the certainty or uncertainty associated with each classifier’s prediction, resulting in a more probabilistic final prediction.
4 Experimental results and discussion
This section discusses the experimental design and results of all experiments to predict the survival of cardiac patients. First, we present the results with a complete feature set, and then present the results with an important feature set. We then discuss the experiments carried out to evaluate the effectiveness of the proposed ensemble technique. In the end, this section compares the proposed approach to the existing approach.
4.1 Experimental setup
The experimental results were implemented in the Python environment using various libraries. Experiments are carried out on an Intel ® Core ™ i5-4300U @ 2.50 GHz processor with 8 GB of RAM running Windows 7 to evaluate the performance of the algorithms.
4.2 Evaluation Metrics
To assess the performance and effectiveness of the proposed model, various evaluation indicators are used, such as sensitivity, accuracy, precision, f1-score, negative predictive value (NPV) and Matthews correlation coefficient (MCC). These metrics are calculated using a confusion matrix. The confusion matrix helps to calculate all four indicators. These definitions are as follows:
-
TP = True Positive (Correctly Identified Positive Class).
-
TN = True Negative (Correctly Identified Negative Class).
-
FP = False Positive (Incorrectly Identified Positive Class).
-
FN = False Negative (Incorrectly Identified Positive Class).
Taking into account the counts of TP, TN, FP, and FN, the following performance evaluation metrics are calculated:
In addition, another metric, area under the curve (AUC), was also used in this study to distinguish between classes. The higher the AUC, the more efficient the model is in distinguishing between positive and negative classes.
4.3 Results of feature selection processes
Table 3 shows the features selected by RF, Boruta and LightGBM feature selection methods. We used six feature selection techniques. Using these techniques, we reduced the dataset and selected the seven most important features from 11 attributes for diagnosing heart disease. The selected features will help doctors understand the significance of the differences in the selected features. The most important features selected by Pearson Correlation are shown in Fig. 3. The most important features predicting heart disease are St slop, depression, and exercise-induced angina. According to the findings, the ranking scores for these features are 0.517, 0404, and 0.484, respectively. We used six different feature selection methods to estimate whether different features would be selected by different methods. For this, we used inspection methods to find the most frequent attributes across the features. This process is mentioned in Table 4. We tick (√) the most frequently occurring features. In Table 4, we finally obtained 7 features from analysis. Now, these 7 (+ 1 target) features will be used for further analysis, and a comparative study with all the features will be performed to obtain the improved accuracy of the classifier.
The highest rankings are age, cholesterol, heart rate, depression, blood pressure, exercise-induced angina and St slope.
4.4 Comparison of various ML algorithms on different features
A comparative analysis of ML classifiers was performed on various input features of the heart disease dataset. First, nine ML classifiers were applied to all features of the heart disease dataset. Second, six feature selection methods were implemented to extract some of the relevant features, and the same nine ML classifiers were applied again to conduct a comparative study. Some classifiers performed well on the evaluation metrics, but some classifiers performed poorly. The three best classifiers that perform well on the selected features are selected to propose a new ensemble classification method.
4.4.1 Classifiers performance comparison based on accuracy
The accuracy rate can be obtained by dividing the number of all correct predictions by the total number of predictions. Figure 4 shows the accuracy of the performance of different types of classifiers on a data set with 11 features and a data subset with 7 features. The most accurate prediction is 94.46% obtained from XGBoost, while the accuracy of KNN using 11 features is 93.19%. The accuracy of GBM and RF are very similar (94.04%). The RF accuracy result showed an improvement of 1.27% with 7 significant features and achieved an accuracy of 95.31%. XGB showed a significant improvement using 7 important features and showed a 1.28% improvement in accuracy and reached an accuracy of 95.74%. The NB Classifier produces the lowest accuracy of 79.57%. The GBM, KNN, MLP, and AdaBoost classifiers provide 94.46%, 92.76%, 84.68%, and 86.8% accuracy, respectively.
4.4.2 Classifiers performance comparison based on sensitivity
Recognizing the authenticity of all relevant instances is the complete ability of the model. The sensitivity score is an important performance matrix because it is very important to accurately classify patients with heart disease. Figure 5 shows the sensitivity scores of the various classifiers using the full set of features and the seven key features. Considering 13 features, the highest sensitivity score of 94.3% was obtained from KNN and RF Classifier. We obtained 90.24%, 92.68%, 92.68%, 84.55%, 85.36%, 84.55%, 83.73% sensitivity scores of DTC, XGB, GBM, MLP, LR, AdaBoost, and NB classifiers respectively. XGB sensitivity improved by 5.69% with seven key features, achieving 98.37% sensitivity. GBM has shown significant improvements using seven key features, with a 4% increase in sensitivity and a 96.74% sensitivity. A very low sensitivity score of 82.92% was obtained with the NB algorithm. All ML classifiers except NB and DTC have excellent performance on 7 selected features, rather than a complete feature set.
4.4.3 Classifiers performance comparison based on precision
Precision is the ratio between true positives and all positives. For our problem statement, precision is also used to evaluate the performance of the classifier. The XGB model gave outstanding results of over 96%, and the NB model had the lowest accuracy score of 86.55% using the 11 features of the dataset. When applied to seven selected features, the DTC gave the highest precision (94.82%) and the NB classifier gave the lowest precision (79%). We obtained 91.4%, 93.75%, 93.79%, 92.96%, 83.72%, 82.22%, 83.82% precision scores of KNN, RF, XGB, GBM, MLP, LR, and AdaBoost classifiers respectively. Figure 6 shows a performance comparison of the precision of a full set of features with seven key features.
4.4.4 Classifiers performance comparison based on F1-score
F-score is a way to combine the precision and recall of the model, and is defined as the harmonic average of Precision and Recall. The F1 score reaches the highest value at 1 and the lowest value at 0. For 11 features, XGB achieved the highest F1 score of 94.6%. NB has the lowest F1 score for 11 features (85.12%), and the results of KNN, RF, DTC, and GBM classifiers are 93.54%, 94.3%, 90.61%, and 94.21%, respectively. After reducing the number of features, the F1 score increased. For seven features, the XGB score was the highest, and most other classifiers also had better F1 score. Considering 7 important features, the highest F1 score was obtained with the XGB model, which gives the highest f1-score of 96%, and RF gives the second highest score, which is exactly 95.6%. Other classifiers such as KNN, DTC, GBM, MLP, LR, and AdaBoost obtained 93.22%, 92.05%, 94.82%, 85.71%, 86.04%, and 88.03% of f1-score respectively. The f1 score of the NB model is the lowest at 80.95%. Figure 7 compares the performance of all classifiers in terms of F1 scores.
4.4.5 Classifiers performance comparison based on MCC
MCC measures the correlation between the true category and the predicted label. The higher the correlation between the true value and the predicted value, the better the prediction. The MCC is a more reliable statistical rate that gives a high score only if the prediction performs well in all four categories of the confusion matrix. The MCC of various algorithms is explained before and after feature selection. The MCC of various classifiers are depicted in Fig. 8. Considering 11 features, the highest MCC of 89% was obtained from XGB Classifier. We obtained 86.35%, 88.05%, 80.39%, 88.12%, 72.86%, 73.67%, 73.76%, 69.38% MCC of KNN, RF, DTC, GBM, MLP, LR, AdaBoost, and NB classifiers respectively. After decreasing the number of features, the MCC increased. For 7 features, the highest MCC of 91.57% is achieved with XGB, which outperforms all other algorithms. RF provides the second highest MCC which is exactly 90.68%.
4.4.6 Classifiers performance comparison based on NPV
NPV is the percentage of test-negative patients who do not have the disease. Figure 9 shows the NPV of different algorithms and feature sets. The NB algorithm achieved a very poor NPV of 82.76%, while RF achieved the highest NPV of 93.75% when applied to the original 11 features. The NPV of KNN, XGB, and GBM based on 11 features are 93.64%, 92.31%, and 92.17%, respectively. For seven features, the best NPV of 98.11% was achieved using XGB, which is superior to all other algorithms. RF provides the second highest NPV, which is exactly 97.2%. Other classifiers such as KNN, DTC, GBM, MLP, LR, and AdaBoost obtained 94.39%, 89.08%, 90.27%, 87.62%, 88%, and 90.91% of NPV respectively.
4.4.7 Classifiers performance comparison based on AUC
The AUC curve is a performance measure of a classification problem with various threshold settings. It is usually used to measure the quality of the classification model; it is a probability curve that plots the true positive rate (TPR) and false positive rate (FPR). The outcomes for AUC are depicted in Fig. 10. For 11 features, the highest AUC of 94.55% is achieved with XGB. NB had the lowest AUC for 11 features (84.72%). After reducing the number of features, AUC increases. For 7 features, XGB has the highest AUC (95.61%), and most other classifiers have AUC better than 13 features.
4.5 Performance comparison of the proposed ensemble method with different classifiers
After analyzing the results described above in Sect. 4.4, the three best classifiers are selected based on various evaluation metrics such as RF, XGB, GBM, and the classification results are passed to different voting mechanisms to make the final predictions. To check the effectiveness of the proposed method, a comparative study with other well-known ML methods is carried out. Methods include KNN, RF, DTC, XGB, GBM, MLP, LR, AdaBoost, and NB. Table 5 shows a comparison of the results of the proposed ensemble model with various other ML classifiers. The experimental results demonstrate that the proposed ensemble model outperforms other methods in terms of accuracy, sensitivity, precision, F1-score, NPV, AUC, and MCC. Among the ensemble models developed for prediction, the majority voting technique proves to be highly effective, achieving remarkable performance with 96.17% classification accuracy, 98.37% sensitivity, 94.53% precision, 96.41% F1-score, and 98.13% NPV. The proposed ensemble algorithm significantly enhances the prediction accuracy by utilizing only seven features, surpassing the classification accuracy of all individual classifiers. This improvement is particularly noteworthy in the context of medical diagnostic systems, where high sensitivity is crucial for accurately identifying heart disease patients. The proposed model not only achieves high sensitivity and specificity but also demonstrates superior ability in distinguishing disease data with enhanced accuracy. In summary, the experimental results highlight the superiority of the proposed ensemble model in terms of its comprehensive performance metrics, particularly in the heart disease diagnosis where accurate disease identification is of utmost importance. The model’s ability to leverage a minimal set of features while achieving superior classification accuracy makes it a promising approach for effective and efficient heart disease diagnosis. The confusion matrix of the proposed ensemble model, specifically the Majority Voting and Soft Voting techniques, is illustrated in Fig. 11. It reveals that the Majority Voting ensemble method achieves a relatively low misclassification rate. Out of a total of 235 cases, the Majority Voting ensemble successfully classifies 226 samples. This implies that the ensemble model accurately assigns these samples to their respective classes based on the majority decision from the individual classifiers. By aggregating the predictions of multiple classifiers, the ensemble model effectively reduces errors and enhances the overall classification performance. The high success rate of 226 out of 235 samples demonstrates the robustness and efficacy of the Majority Voting ensemble method. It showcases the model’s ability to make accurate predictions by leveraging the collective knowledge and diverse perspectives of the individual classifiers. The Majority Voting ensemble method minimizes the impact of outliers or individual classifier biases, resulting in improved classification accuracy and reliable predictions.
4.6 Comparative analysis with previously reported methods
In this section, the experimental results of the proposed method are compared with the experimental results of other methods described in the literature. Performance of the proposed method in terms of accuracy compared to existing methods in the heart disease diagnosis literature. Thus, Table 6 summarizes the predictive accuracy of our proposed method with other well-performing methods proposed in the recent literature. The table presents a summary of our proposed method performance compared to other relevant studies. Table 6 demonstrates the excellent performance of the proposed ensemble model compared to previous studies. The results highlight the model’s ability to accurately diagnose heart disease, making it a valuable tool in the field of healthcare. The robust performance of the proposed ensemble model, as showcased by its high accuracy, surpasses the results reported in prior research. Previous models for identifying and predicting heart failure disease have not achieved the same level of accuracy. However, the newly proposed ensemble model improved predictive accuracy by utilizing only 7 features and outperformed existing techniques in terms of classification accuracy. This indicates the model’s superiority in accurately identifying and classifying heart disease cases. Furthermore, the proposed method offers practical advantages in terms of implementation within healthcare organizations. Its effectiveness and accuracy make it a reliable diagnostic system that can be readily integrated into existing healthcare infrastructures. The ease of implementation ensures that healthcare professionals can easily adopt and utilize the proposed system for efficient and accurate diagnosis of heart disease.
5 Conclusion
Heart disease is very complicated and causes many deaths every year. If the early symptoms of heart disease are ignored, patients may have serious health problems. Early diagnosis helps prevent the progression of the disease. In this study, we proposed an improved ensemble-based model for detecting heart disease. We reported the performance of nine ML classifiers on a full dataset using various performance metrics such as accuracy, precision, sensitivity, F1-score, NPV, MCC, AUC. We used six feature selection techniques and select 7 features based on feature importance and rank. The comparison between the ensemble method and nine classifiers (KNN, RF, DTC, XGB, GBM, MLP, LR, AdaBoost, and NB) is applied to the full set of features and selected features in the heart disease dataset. Compared to other ML methods and recent scientific work, the proposed ensemble classifier shows better performance and is able to detect heart disease with an accuracy of 96.17% and a sensitivity of 98.37%. We also know that irrelevant features also reduce the performance of the diagnostic system. Therefore, another innovation of our study was to use a feature selection algorithm to select the appropriate features which improved the performance of the diagnostic system. This work has the potential to improve the healthcare system, and can be used to predict heart disease risks and effectively assist clinical consultations.
References
Sanz, M., Marco del Castillo, A., Jepsen, S., Gonzalez-Juanatey, J.R., D’Aiuto, F., Bouchard, P., Wimmer, G.: Periodontitis and cardiovascular diseases: Consensus report. J. Clin. Periodontol. 47(3), 268–288 (2020)
Allen, L.A., Stevenson, L.W., Grady, K.L., Goldstein, N.E., Matlock, D.D., Arnold, R.M., Spertus, J.A.: Decision making in advanced heart failure: A scientific statement from the American Heart Association. Circulation. 125(15), 1928–1952 (2012)
Pouriyeh, S., Vahid, S., Sannino, G., De Pietro, G., Arabnia, H., Gutierrez, J.: A comprehensive investigation and comparison of machine learning techniques in the domain of heart disease. In 2017 IEEE symposium on computers and communications (ISCC), pp. 204–207. IEEE. July 2017
Ghwanmeh, S., Mohammad, A., Al-Ibrahim, A.: Innovative artificial neural networks-based decision support system for heart diseases diagnosis. J. Intell. Learn. Syst. Appl. 5(3), 176–83 (2013)
Sevakula, R.K., Verma, N.K.: Assessing generalization ability of majority vote point classifiers. IEEE Trans. neural networks Learn. Syst. 28(12), 2985–2997 (2016)
Li, H., Cui, Y., Liu, Y., Li, W., Shi, Y., Fang, C., Lu, Y.: Ensemble learning for overall power conversion efficiency of the all-organic dye-sensitized solar cells. IEEE Access. 6, 34118–34126 (2018)
Shamrat, F.J.M., Raihan, M.A., Rahman, A.S., Mahmud, I., Akter, R.: An analysis on breast disease prediction using machine learning approaches. Int. J. Sci. Technol. Res. 9(02), 2450–2455 (2020)
Singh, D., Samagh, J.S.: A comprehensive review of heart disease prediction using machine learning. J. Crit. Reviews. 7(12), 281–285 (2020)
Asif, S., Wenhui, Y., Tao, Y., Jinhai, S., Jin, H.: An Ensemble Machine Learning Method for the Prediction of Heart Disease. In 2021 4th International Conference on Artificial Intelligence and Big Data (ICAIBD), pp. 98–103. IEEE. May 2021
Liu, X., Wang, X., Su, Q., Zhang, M., Zhu, Y., Wang, Q., Wang, Q.: A hybrid classification system for heart disease diagnosis based on the RFRS method. Comput. Math. Methods Med. (2017). https://doi.org/10.1155/2017/8272091
Amin, M.S., Chiam, Y.K., Varathan, K.D.: Identification of significant features and data mining techniques in predicting heart disease. Telematics Inform. 36, 82–93 (2019)
Atallah, R., Al-Mousa, A.: Heart disease detection using machine learning majority voting ensemble method. In 2019 2nd international conference on new trends in computing sciences (ictcs), pp. 1–6. IEEE. October, 2019
Kannan, R., Vasanthi, V.: Machine learning algorithms with ROC curve for predicting and diagnosing the heart disease. In: Soft Computing and Medical Bioinformatics, pp. 63–72. Springer, Singapore (2019)
Gudadhe, M., Wankhade, K., Dongre, S.: Decision support system for heart disease based on support vector machine and artificial neural network. In 2010 International Conference on Computer and Communication Technology (ICCCT), pp. 741–745. IEEE, September, 2010
Prasad, R., Anjali, P., Adil, S., Deepa, N.: Heart disease prediction using logistic regression algorithm using machine learning. Int. J. Eng. Adv. Technol. 8(3S), 659–662 (2019)
Melillo, P., De Luca, N., Bracale, M., Pecchia, L.: Classification tree for risk assessment in patients suffering from congestive heart failure via long-term heart rate variability. IEEE J. biomedical health Inf. 17(3), 727–733 (2013)
Nalluri, S., Saraswathi, V., Ramasubbareddy, R., Govinda, S., K., Swetha, E.: Chronic heart disease prediction using data mining techniques. In: Data Engineering and Communication Technology, pp. 903–912. Springer, Singapore (2020)
Sapra, L., Sandhu, J.K., Goyal, N.: Intelligent method for detection of coronary artery disease with ensemble approach. In: Advances in Communication and Computational Technology, pp. 1033–1042. Springer, Singapore (2021)
Raza, K.: Improving the prediction accuracy of heart disease with ensemble learning and majority voting rule. In: U-Healthcare Monitoring Systems, pp. 179–196. Academic Press, Cambridge (2019)
Mohan, S., Thirumalai, C., Srivastava, G.: Effective heart disease prediction using hybrid machine learning techniques. IEEE Access 7, 81542–81554 (2019)
Zomorodi-moghadam, M., Abdar, M., Davarzani, Z., Zhou, X., Pławiak, P., Acharya, U.R.: Hybrid particle swarm optimization for rule discovery in the diagnosis of coronary artery disease. Expert Syst. 38(1), e12485 (2021)
Geweid, G.G., Abdallah, M.A.: A new automatic identification method of heart failure using improved support vector machine based on duality optimization technique. IEEE Access 7, 149595–149611 (2019)
Haq, A.U., Li, J.P., Memon, M.H., Nazir, S., Sun, R.: A hybrid intelligent system framework for the prediction of heart disease using machine learning algorithms. Mob. Inform. Syst. (2018). https://doi.org/10.1155/2018/3860146
Rashmi, G.O., Kumar, U.M.A.: Machine learning methods for heart disease prediction. Int. J. Eng. Adv. Technol. 8(5S), 220–223 (2019)
Sharma, S., Parmar, M.: Heart diseases prediction using deep learning neural network model. Int. J. Innovative Technol. Exploring Eng. (IJITEE). 9(3), 2244–2248 (2020)
Dwivedi, A.K.: Performance evaluation of different machine learning techniques for prediction of heart disease. Neural Comput. Appl. 29(10), 685–693 (2018)
Alizadehsani, R., Habibi, J., Hosseini, M.J., Mashayekhi, H., Boghrati, R., Ghandeharioun, A., Sani, Z.A.: A data mining approach for diagnosis of coronary artery disease. Comput. Methods Programs Biomed. 111(1), 52–61 (2013)
Guidi, G., Pettenati, M.C., Melillo, P., Iadanza, E.: A machine learning system to improve heart failure patient assistance. IEEE J. Biomed. Health Inform. 18(6), 1750–1756 (2014)
Abdar, M., Acharya, U.R., Sarrafzadegan, N., Makarenkov, V.: NE-nu-SVC: A new nested ensemble clinical decision support system for effective diagnosis of coronary artery disease. IEEE Access. 7, 167605–167620 (2019)
Qin, C.J., Guan, Q., Wang, X.P.: Application of ensemble algorithm integrating multiple criteria feature selection in coronary heart disease detection. Biomed. Eng. 29(06), 1750043 (2017)
Abdar, M., Książek, W., Acharya, U.R., Tan, R.S., Makarenkov, V., Pławiak, P.: A new machine learning technique for an accurate diagnosis of coronary artery disease. Comput. Methods Programs Biomed. 179, 104992 (2019)
Shah, D., Patel, S., Bharti, S.K.: Heart disease prediction using machine learning techniques. SN Comput. Sci. 1(6), 1–6 (2020)
Latha, C.B.C., Jeeva, S.C.: Improving the accuracy of prediction of heart disease risk based on ensemble classification techniques. Inf. Med. Unlocked. 16, 100203 (2019)
Nasarian, E., Abdar, M., Fahami, M.A., Alizadehsani, R., Hussain, S., Basiri, M.E., Sarrafzadegan, N.: Association between work-related features and coronary artery disease: a heterogeneous hybrid feature selection integrated with balancing approach. Pattern Recognit. Lett. 133, 33–40 (2020)
Dua, D., Graff, C.: UCI Machine Learning Repository. University of California, School of Information and Computer Science, Irvine, CA (2019)
Alizadehsani, R., Roshanzamir, M., Abdar, M., Beykikhoshk, A., Khosravi, A., Panahiazar, M., Sarrafzadegan, N.: A database for using machine learning and data mining techniques for coronary artery disease diagnosis. Sci. Data. 6(1), 1–13 (2019)
Kursa, M.B., Rudnicki, W.R.: Feature selection with the Boruta package. J. Stat. Softw. 36, 1–13 (2010)
Bashir, S., Qamar, U., Khan, F.H.: A multicriteria weighted vote-based classifier ensemble for heart disease prediction. Comput. Intell. 32(4), 615–645 (2016)
Ali, L.I., Niamat, A., Golilarz, N.A., Ali, A., Xingzhong, X.: An expert system based on optimized stacked support vector machines for effective diagnosis of heart disease. IEEE Access 4, 2169–3536 (2019)
Paul, A.K., Shill, P.C., Rabin, M., Islam, R., Murase, K.: Adaptive weighted fuzzy rule-based system for the risk level assessment of heart disease. Appl. Intell. 48(7), 1739–1756 (2018)
Dinesh, K.G., Arumugaraj, K., Santhosh, K.D., Mareeswari, V.: ‘Prediction of cardiovascular disease using machine learning algorithms, In: Proceedings International Conference on Current Trends towards Converging Technologies (ICCTCT), Coimbatore, India, pp. 1–7 (2018)
Funding
This work was supported in part by the National Key Research and Development Program of China under Grant 2019YFA0706400 and Grant 2019YFA0706402, in part by the Pre-Research Funds for Equipment of China under Grant 61409220115.
Author information
Authors and Affiliations
Contributions
Sohaib Asif: Data curation, Methodology, Validation, Writing – original draft. Wenhui Yi: Conceptualization, Investigation, Supervision, Writing – review & editing. Jin Hou and Jinhai Si: Formal analysis, Validation, Investigation. Qurrat ul Ain and Yueyang Yi: Analysis, Validation, Writing – review & editing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
For this type of study, formal consent is not required.
Experiments involving human and/or animal participants
This paper does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Asif, S., Wenhui, Y., ul Ain, Q. et al. Improving the accuracy of diagnosing and predicting coronary heart disease using ensemble method and feature selection techniques. Cluster Comput 27, 1927–1946 (2024). https://doi.org/10.1007/s10586-023-04062-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10586-023-04062-2