Abstract
The subfamily Arvicolinae consists of a great number of species with highly diversified karyotypes. In spite of the wide use of arvicolines in biological and medicine studies, the data on their karyotype structures are limited. Here, we made a set of painting probes from flow-sorted chromosomes of a male Palearctic collared lemming (Dicrostonyx torquatus, DTO). Together with the sets of painting probes made previously from the field vole (Microtus agrestis, MAG) and golden hamster (Mesocricetus auratus, MAU), we carried out a reciprocal chromosome painting between these three species. The three sets of probes were further hybridized onto the chromosomes of the Eurasian water vole (Arvicola amphibius) and northern red-backed vole (Myodes rutilus). We defined the diploid chromosome number in D. torquatus karyotype as 2n = 45 + Bs and showed that the system of sex chromosomes is X1X2Y1. The probes developed here provide a genomic tool-kit, which will help to investigate the evolutionary biology of the Arvicolinae rodents. Our results show that the syntenic association MAG1/17 is present not only in Arvicolinae but also in some species of Cricetinae; and thus, should not be considered as a cytogenetic signature for Arvicolinae. Although cytogenetic signature markers for the genera have not yet been found, our data provides insight into the likely ancestral karyotype of Arvicolinae. We conclude that the karyotypes of modern voles could have evolved from a common ancestral arvicoline karyotype (AAK) with 2n = 56 mainly by centric fusions and fissions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The subfamily Arvicolinae includes voles, muskrats, and lemmings and is recognized as the most speciose subfamily in mammals. This group of rodents is characterized by a great diversity and distribution of species with a strikingly similar morphology. This has led to the question as to whether chromosomal variation could underlie its rapid speciation. Intensive karyotype investigations of Arvicolinae, which started in the 1960s (i.e., soon after the conventional methods of cytogenetics were developed), have revealed in this group one of the highest interspecific variation among vertebrates (Modi 1987a, b).
Indeed, the diploid chromosome numbers (2n) in voles and lemmings vary from 17 to 86 (Chernyavsky and Kozlovsky 1980; Modi 1987b). Pericentric inversions, both autosomal and sex chromosomal translocations, heteromorphism between homologous chromosome pairs, and Robertsonian polymorphisms have been identified in various arvicoline species. Such rapid and radical changes in karyotype pose a challenge to comparative analyses based on chromosome banding patterns alone and necessitate the use of molecular cytogenetic tools.
The first study devoted to the comparison of Arvicolinae karyotypes by chromosome painting was published in 2006 (Li et al. 2006) followed by four further studies (Romanenko et al. 2007; Sitnikova et al. 2007; Lemskaya et al. 2010; Bakloushinskaya et al. 2012). To date, genome-wide homology maps have been established for the following Arvicolinae species: Eothenomys militus, E. proditor, Neodon (Microtus) clarkei (Li et al. 2006), Alexandromys oeconomus, M. agrestis (Sitnikova et al. 2007), Ellobius talpinus, E. lutescens (Romanenko et al. 2007), E. tancrei (Bakloushinskaya et al. 2012), and eight species from the Arvicolini tribe (subtaxon of Arvicolinae)–Microtus arvalis, M. rossiaemeridionalis, M. socialis, M. dogramacii, M. guentheri, Terricola daghestanicus, Lasiopodomys gregalis, and Alexandromys maximowiczii (Lemskaya et al. 2010).
Despite the long-lasting interests in the evolution of arvicolines many important questions concerning relationships within this taxon remain unclear. Several attempts have been made to solve phylogenetic relationships in Arvicolinae using molecular methods (e.g., DeWoody et al. 1999; Mazurok et al. 2001; Jaarola et al. 2004; Triant and Dewoody 2006; Bannikova et al. 2010; Yannic et al. 2012). Estimates of the divergence of the extant genera in Arvicolinae vary from about 3–5 million years ago (Chaline and Graf 1988) to 5–9 (Repenning 1990; Conroy and Cook 1999). More recent data are also variable and depend on the number and type of sequences included in the analysis. Thus, the basal radiation for genera Dicrostonyx, Prometheomys, Onatra and tribe Lemmini was estimated as 6.4–7.2, 6.8–7.7, 7.7–9.2, and 7.2–8.3 million years ago (mya), respectively (Abramson et al. 2009b), while the divergence of the most modern genera took place from 3.5 to 1 mya (Abramson et al. 2009b; Bannikova et al. 2010). In the third edition of “Mammal species of the world”, the subfamily Arvicolinae was subdivided into 12 tribes, 28 genera, and 151 species (Carleton and Musser 2005). However, many groups include subspecies with potential species status. It is expected that the number of the recognized species in the group will increase with more detailed molecular and cytogenetic investigations (e.g., Yannic et al. 2012). Recently, the taxonomy of Arvicolinae was revised substantially based on new molecular data (Abramson et al. 2011; Pavlinov and Lissovsky 2012).
The Palearctic collared lemming (Dicrostonyx torquatus) belongs to a group of species representing a basal branch of Arvicolinae. The karyotype of the Palearctic collared lemming has been in the focus of cytogenetic studies because of its unusual system of sex chromosomes (XmXm, XmXf, XfO for females, and XmO for males, Gileva and Chebotar 1979; Gileva 1980; Fredga 1983) and variable number of additional (supernumerary) B chromosomes. Moreover, the karyotyping of many animals from different populations has shown that their diploid autosomal numbers were also variable (Gileva 1980). Here, we made a new set of painting probes from the Palearctic collared lemming to clarify the species karyotype. The detailed investigation of the D. torquatus karyotype not only identifies the particulars of arvicoline phylogeny, but also resolves many questions concerning the origin and evolution of sex chromosomes and B chromosomes.
We have carried out cross-species chromosome painting with the newly made probes from D. torquatus to establish chromosome homologies between the three vole species belonging to different tribes: Arvicola amphibious, Arvicolini; D. torquatus, Dicrostonychini; and Myodes rutilus, Myodini. All species studied have a wide distribution in Eurasia, have been successfully bred in the laboratory, and have formed colonies. A. amphibius and M. rutilus are widely used in research on physiology, parasitology, etc. Although the data on the structure of their karyotypes are limited, some data are available on the pairing of sex chromosomes and the distribution of repeats (Borodin et al. 1995; Acosta et al. 2010; de la Fuente et al. 2012).
To facilitate karyotype comparison of A. amphibius, D. torquatus and M. rutilus and to link it to previous studies, we also used paints from the field vole (Microtus agrestis, 2n = 50, Sitnikova et al. 2007) and the golden hamster (Mesocricetus auratus, 2n = 44, Romanenko et al. 2006). We have integrated our results with previously published comparative chromosome maps of the Arvicolinae species using D. torquatus painting probes for the additional A. oeconomus and M. agrestis karyotype investigations (Romanenko et al. 2007; Sitnikova et al. 2007; Lemskaya et al. 2010; Bakloushinskaya et al. 2012).
Materials and methods
Species used in this study are listed in Table 1. Chromosome suspensions from early passages of primary fibroblast cell lines of all species were obtained from the Laboratory of Animal Cytogenetics, Institute of Molecular and Cellular Biology, Russia, and made following Sitnikova et al. 2007. Fibroblast cell lines were established as previously published (Sitnikova et al. 2007). Each suspension was tested for chromosomal rearrangements by conventional cytogenetic analysis. Each cell line had a stable karyotype.
The sets of paints derived from flow-sorted chromosomes of the field vole and the golden hamster have been described previously (Yang et al. 1995; Romanenko et al. 2006). The Palearctic collared lemming paints were generated at the Cambridge Resource Centre for Comparative Genomics, by DOP-PCR amplification of flow-sorted chromosomes and labeled with biotin and digoxigenin-dUTPs (Medigen) by DOP-PCR amplification (Telenius et al. 1992; Yang et al. 1995). The probe specific for the short p-arm of the X chromosome of the Palearctic collared lemming was made by microdissection as previously described (Weimer et al. 2000).
GTG-banding on all metaphase chromosomes prior to fluorescent in situ hybridization (FISH) was performed using trypsin treatment (Seabright 1971). A schematic representation of the cross-species painting experiments is shown in Fig. 1. FISH was performed according to previously published protocols (Yang et al. 1999; Graphodatsky et al. 2000).
A schematic representation of the comparative chromosome painting strategies followed with the Palearctic collared lemming (D. torquatus), field vole (M. agrestis, MAG), golden hamster (M. auratus, MAU), Eurasian water vole (A. amphibius), northern red-backed vole (M. rutilus), and root vole (A. oeconomus). Arrow tails indicate species (bold type) whose chromosome-specific painting probes were used. Arrow heads indicate species whose metaphases were used to detect homologous regions. Grey arrows mark experiments that were made earlier (Sitnikova et al. 2007)
The telomeric DNA probe was made by PCR using the oligonucleotides (TTAGGG)5 and (CCCTAA)5 (Ijdo et al. 1991). Clones of human ribosomal DNA containing the complete 18S-rRNA and 28S-rRNA genes were obtained as described (Maden et al. 1987) and labeled by biotin and/or digoxigenin-dUTPs (Medigen).
Results
Karyotype description of D. torquatus
A fibroblast cell line derived from a male animal specimen collected from the Vaygach Island (Arctic Sea) was karyotyped. We noticed a variation in the chromosome number between cells due to the presence of mitotically unstable additional (B) chromosomes. The largest chromosome of the complement was a single metacentric accompanied by a number of smaller acrocentrics, submetacentrics, and metacentrics, identifiable only by differential staining. Analysis of high-resolution G-banded karyotypes revealed a complex sex chromosome system consisting of three partially homologous chromosomes–the largest metacentric, the largest submetacentric, and a small acrocentric (Fig. 2). The atypical large X chromosome in this species is a result of a translocation of the ancestral X chromosome onto an autosome, and is the largest chromosome in the D. torquatus karyotype. According to the arm homology and the standard nomenclature, the sex chromosome system should be defined as X1X2Y1, where the giant metacentric X1 includes the evolutionary old “X” part (the p-arm) and the Xq-arm is homologous to the long arm of Y1, the largest submetacentric in the DTO karyotype. The short arm of Y1 is homologous to acrocentric X2. Therefore, the diploid number of this male specimen is 2n = 45, including 21 autosome pairs and 3 sex chromosomes, with the numbers of B chromosomes varying from 8 to 14 in different cells.
GTG-banded karyotype of D. torquatus. Black squares mark the position of a centromere. Vertical black bars mark the localization of chromosomal segments of M. agrestis (MAG), M. auratus (MAU). Numbers along the vertical lines correspond to the chromosome numbers of M. agrestis and M. auratus. Black triangles indicate sites of localization of rDNA clusters, grey triangles indicate localization of the largest interstitial telomeric block (shown for chromosome 1 only)
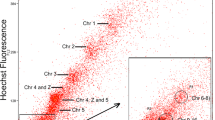
The flow karyotype of D. torquatus
The chromosome complement of D. torquatus was resolved into 29 separate peaks by flow cytometry (Fig. 3). The chromosomal content of each peak was identified by hybridizing the probes derived from each peak onto GTG-banded D. torquatus chromosomes (data not shown). Eighteen single chromosome-specific paining probes were obtained (1–8, 13–19, Х1, X2, and Y1). Six peaks gave signals on B chromosomes only and were not used in this study. Two probes each painted two pairs of autosomes (6 + 9, 10 + 12) and three peaks contained a mixture of one autosome and B chromosomes (11 + Bs, 20 + Bs, and 21 + Bs). FISH with different sets of painting probes (see succeeding sentences) allowed us to distinguish four A chromosomes of two pairs (Nos. 20–21 in the karyotype) from a group of small B chromosomes. The two largest chromosomes (i.e., X1 and Y1) of D. torquatus were each resolved into separate peaks, but their flow peaks sort too far away from the remaining chromosomes and are not shown in Fig. 3. In total, 23 painting probes, containing all autosomes, X1, X1, and Y1 chromosomes of D. torquatus, were used in this study. FISH experiments did not reveal any signal potentially corresponding to a “true” Y-chromosome or pseudoautosomal region.
Reciprocal chromosome painting between D. torquatus and M. agrestis
The 21 M. agrestis (MAG) autosomal probes revealed 33 conserved segments in the D. torquatus genome (Fig. 2, Supplementary Material 1). Examples of fluorescent in situ hybridizations are shown in Fig. 4. The MAG Y probe did not produce any specific signal on any chromosome in the D. torquatus genome. The MAG X probe painted only the p-arm of the D. torquatus X1 chromosome. The 23 D. torquatus (DTO) autosomal probes revealed 33 conserved segments in the M. agrestis genome (Fig. 5, Supplementary Material 1). The probe DTOX1p painted the M. agrestis X chromosome.
Examples of fluorescent in situ hybridization: a MAG12 (green) and MAG18 (red) onto D. torquatus chromosomes, b MAG24 (green) and MAG17 + 12 (red) onto D. torquatus chromosomes, c DTO2 (green) and DTO19 (red) onto M. agrestis chromosomes, d DTO5 (green) and DTO2 (red) onto M. agrestis chromosomes, e DTO13 (green) and DTO6 + 9 (red) onto A. amphibius chromosomes, and f MAG19 + 20 (green) and MAG13 + 14 (red) onto C. rutilus chromosomes. GTG-banded chromosomes are shown on the right. Scale bar is 10 μm
GTG-banded karyotype of M. agrestis. Black squares mark the position of a centromere. Vertical black bars mark the localization of chromosomal segments of D. torquatus (DTO) and M. auratus (MAU). Numbers beside the vertical lines correspond to chromosomes of D. torquatus and M. auratus. Black triangles indicate the localization of rDNA clusters
Reciprocal chromosome painting between D. torquatus and M. auratus
Examples of fluorescence in situ hybridizations are shown in Fig. 4. The 21 M. auratus paints (MAU) delineated 44 homologous autosomal segments in the D. torquatus genome (Fig. 2, Supplementary Material 1). The MAU X probe only hybridized to the p-arm of the D. torquatus X1 chromosome. The 23 D. torquatus (DTO) autosomal probes revealed 43 conserved segments in the M. auratus genome (Fig. 6, Supplementary Material 1).
GTG-banded karyotype of M. auratus. Black squares mark the position of a centromere. Vertical black bars mark the localization of chromosomal segments of M. agrestis (MAG), D. torquatus (DTO), and M. musculus (MMU, Romanenko et al. 2007). Numbers beside the vertical lines correspond to chromosomes of M. agrestis and D. torquatus. Black triangles–localization of rDNA clusters, grey–telomeric blocks (showing the brightest signals only)
Reciprocal chromosome painting between M. agrestis and M. auratus
Examples of fluorescence in situ hybridizations are shown in Fig. 4. The 21 M. agrestis autosomal probes revealed 40 conserved segments in the M. auratus genome (Fig. 6, Supplementary Material 1). Reciprocal painting with 21 M. auratus paints delineated 42 homologous autosomal segments in the M. agrestis genome (Fig. 5). The MAU X probe only hybridized to the X chromosome of M. agrestis.
Chromosome painting of A. amphibius, M. rutilus, and A. oeconomus using D. torquatus paints
The results of hybridization in these three species, including the number of hybridization signals produced by each D. torquatus paints per haploid karyotype of the target species and the number of conserved segments, are listed in Supplementary Material 1.
Chromosome painting of A. amphibius and M. rutilus using M. agrestis paints
The results of hybridization of M. agrestis are shown in Supplementary Material 1 as the number of hybridization signals produced by each of the M. agrestis paints per haploid karyotype of the target species and the total number of conserved segments. Most of the large homologous segments defined by chromosome painting also showed conserved banding patterns (Figs. 2, 5, 7–9).
GTG-banded karyotype of A. amphibius. Black squares mark the position of a centromere. Vertical black bars mark the localization of chromosomal segments of M. agrestis (MAG), D. torquatus (DTO), and M. auratus (MAU). Numbers beside the vertical lines correspond to chromosomes of M. agrestis, D. torquatus, and M. auratus. The probes, which were not checked by FISH, are shown by grey lines. Black triangles–localization of rDNA clusters, grey–the largest interstitial telomeric block
GTG-banded karyotype of M. rutilus. Black squares mark the position of a centromere. Vertical black bars mark the localization of chromosomal segments of M. agrestis (MAG), D. torquatus (DTO), and M. auratus (MAU). Numbers beside the vertical lines correspond to chromosomes of M. agrestis, D. torquatus, and M. auratus. Those probes, which were not directly checked by FISH, are shown by grey lines
GTG-banded karyotype of M. oeconomus. Black squares mark the centromeric positions. Vertical black bars mark the localization of chromosomal segments of M. agrestis (MAG), D. torquatus (DTO), M. auratus (MAU), and M. musculus (MMU, Sitnikova et al. 2007). Numbers beside the vertical lines correspond to chromosomes of M. agrestis, D. torquatus, and M. auratus. The M. agrestis and M. auratus probes, signals from which have not detected earlier, are marked in grey (Sitnikova et al. 2007; Lemskaya et al. 2010). Those probes, which were not checked by FISH on D. torquatus, are shown by grey lines. Black triangles mark the localization of rDNA clusters
Precise homology between some regions detected by the same M. agrestis probe in karyotypes of different species (e.g., MAG1 onto MRU3, 9 (Fig. 8) and DTO4, 5, 8 (Fig. 2) could not be established. To resolve ambiguities and to define homologies between karyotypes precisely, selected paints of M. auratus were hybridized to M. rutilus and A. amphibius chromosomes.
Chromosome painting of A. amphibius and M. rutilus using M. auratus paints
All paints of M. auratus were hybridized to A. amphibius chromosomes (Fig. 7). In total, 40 autosomal conserved segments were detected. All paints of M. auratus were hybridized to M. rutilus chromosomes (Fig. 8). In total, 45 autosomal conserved segments were detected.
Distribution of telomeric sites and rDNA clusters
We also investigated distribution of telomeric sites and ribosomal DNA (rDNA) clusters with telomeric DNA and rDNA probes hybridization (see Supplementary Material 2 for detail).
Discussion
The extraordinary karyotype of D. torquatus
A distinctive feature of the genus of the collared lemming, Dicrostonyx, is its wide karyotypic variability, especially in comparison with their closest relatives–the true lemmings (Lemmus). The diploid chromosome numbers in Dicrostonyx vary from 2n = 28 in D. vinogradovi to 2n = 86 in D. torquatus (Chernyavsky and Kozlovsky 1980; Gileva 1983; Orlov and Bulatova 1983). This is in sharp contrast to the uniform diploid number (2n = 50) shared by the four Lemmus species karyotyped so far (Gileva 1983). It was proposed that the variation in diploid numbers in Dicrostonyx is mainly caused by centric fusions of A chromosomes (Gileva 1983). An exceptional variation in the number of B chromosomes results in the high chromosome numbers in D. torquatus, where 44 A chromosomes were considered to be regular elements of the species karyotype (2n = 44 + Bs). Karyotypes with 0 to 42 B chromosomes have been reported in populations of Northern Eurasia (Orlov and Bulatova 1983). An increase in the number of B chromosomes eastwards in populations along the Russian Arctic Coast was suggested by Fredga et al. (1999), but such a clear geographic trend was not supported by a subsequent study (Gileva 2004).
However, chromosome sizes do not allow a reliable distinction between the A and B chromosomes and the identification of B chromosomes should be regarded as provisional without molecular cytogenetic analyses. Only after FISH with M. agrestis probes it become possible to determine that the two small chromosome pairs in the lemming karyotype, which are of the same size as B chromosomes, are homologous to MAG24 and therefore belong to the standard complement. We thus postulate that the diploid number of male D. torquatus is 2n = 45 + Bs (Fig. 2). Interestingly, the “true” male specific Y chromosome was not identified in the karyotype of D. torquatus based on meiotic pairing studies (Gileva and Chebotar 1979; Gileva 1980) and also was not visualized by painting probes in our experiments. This could place D. torquatus in a short list of atypical mammalian species that lack a typical Y chromosome (Romanenko and Volobouev 2012). We proposed that the translocation of the male specific Y chromosome onto an autosome followed by the elimination of most parts of the true Y-chromosome took place during the formation of the present karyotype of the species. However, the potential elimination of ancestral Y chromosome and the absence of modern high-resolution meiotic studies have prevented the establishment of the presence of such a Y-specific region in D. torquatus. Further molecular investigation of D. torquatus with probes containing genes from the standard mammalian pseudoautosomal region are required to verify the proposed karyotype composition. Keeping in mind with the apparent lack of a true Y-chromosome, the system of sex chromosomes could be designated at present as X1X2Y1. The three sex chromosomes might be interpreted as the result of a double translocation event resulting in three monobrachial homologs (Fig. 2). Previously, a similar heteromorphic structure was described in D. torquatus torquatus from the Polar Ural (Gileva 1980). Importantly, the G-banding pattern of X2 (= chr. 19) and of the p-arm of Y1 (= chr. 18) reported by Gileva (1980) is different from that in our specimen. We postulate that the heteromorphic systems of sex chromosomes in different populations of D. torquatus could have involved different autosomes.
Insight into the chromosome arrangements involved in Arvicolinae karyotype evolution
The question about the role of chromosomal rearrangements in speciation remains open. It was shown that some types of rearrangements can influence the reproductive function of heterozygous carriers (Romanenko et al. 2012 and references herein).
Previously published data document various chromosomal rearrangements in Arvicolinae (mainly Robertsonian translocations). To summarize all painting results obtained up to now for arvicolines we present a table showing the distribution of shared syntenic segment associations found in karyotypes of voles, lemming, mole vole, and some hamster species studied previously (Supplementary Material 3). The use of different sets of probes for species comparison enabled us to show that the variety between closely related karyotypes is caused not only by fusions and fissions, but also by a great number of intrachromosomal rearrangements including inversions and the emergence of evolutionary new centromeres (ENC). A cladistic analysis of chromosomal characters identified in all arvicolines studied did not give sufficient resolution to resolve the karyotype relationships of Arvicolinae species so far studied by comparative painting due to the shortage of informative chromosomal characters (data not shown).
The karyotypes of voles and lemmings from the Arvicolinae subfamily have been well studied. Although molecular comparative cytogenetic data do not provide any particular phylogenetic markers for this group, they do indicate the major role of intrachromosomal compared to interchromosomal rearrangements in the karyotype evolution of the subfamily. However, to characterize the intrachromosomal rearrangements fine-resolution molecular cytogenetic probes would be required. Even the use of chromosome painting sets with a variety of cytogenetic features here has not been sufficient for the precise identification of intrachromosomal exchanges occurring in Arvicolinae evolution.
The most intriguing cases are those that have rearrangements inside the segment homologous to MAG2/8 prox = DTO1 = MAU6q/10p/8q prox/9q dist (Fig. 10). Multiple inversions inside this segment were detected previously in different Arvicolini karyotypes (Lemskaya et al. 2010). It was also observed that the centromere position in chromosomes homologous to MAG2/8 varies in Microtus species (Lemskaya et al. 2010). Using M. auratus probes we were able to look deeper into the composition of the segment in different arvicoline karyotypes. We assume that both inversions and centromere reposition events could have occurred during species diversification. However, the evolutionary direction of these rearrangements could not be determined from such a limited number of species. Hybridization of BAC-clones or region-specific microdissected probes is needed for the detailed analysis of these intrachromosomal rearrangements.
The synteny MAG2/8 in karyotypes of different Arvicolinae species: AAM, Arvicola amphibius; AOE, Alexandromys oeconomus; MRU, Myodes rutilus; DTO, Dicrostonyx torquatus; MAG, Microtus agrestis. Black squares mark the centromeric positions. Vertical black bars mark the localization of chromosomal segments of M. auratus
Another controversial issue concerns the segment homologous to MAG1/17. It was proposed previously that MAG1/17 was characteristic for all arvicolines (Lemskaya et al. 2010). But here, with the use of D. torquatus painting probes, we found this association in the M. auratus chromosome 16 (Fig. 6). The MAG1/17 is homologous to DTO5. However, the association MAG1/17 found in A. amphibius, M. rutilus, and A. oeconomus karyotypes contains an additional part of MAG1, homologous to DTO4 (Fig. 9). Thus, based on our new findings, we conclude that MAG1/17 could be present in the ancestral arvicoline karyotype even if its precise structure remains undefined. It is now clear that MAG1/17 (= DTO5) is an ancestral chromosome for Arvicolinae and also for the whole Cricetidae family. The syntenic combination MAG1/17(= DTO4/5) might have appeared later in the evolution of arvicolines, probably after the separation of the branch leading to D. torquatus (Fig. 11). A more detailed investigation of arvicoline species is needed in order to find a node on the tree where the MAG1/17 becomes fixed.
Karyotype relationships in Arvicolinae. The branching order on the tree was adapted from the molecular-based phylogenies as reported by Galewski et al. (2006) and Abramson et al. (2009b). Numbers in squares showed diploid chromosome numbers for nodes. Numbers correspond to M. agrestis chromosomes or chromosomal segments. AAK ancestral Arvicolinae karyotype, minus sign fission, plus sign fusion
Differences in the centromere position for segments homologous to MAG3 were previously reported for a number of Microtus species. Our data show that the segment homologous to MAG3 = MAU5/21/16/14 is characteristic for the whole Arvicolinae subfamily (Supplementary Material 3). In spite of the similar order of M. auratus probes (inside the segment homologous to MAG3) a variation in centromere position between species was found. Such differences in the centromere position in the segment could be caused by a paracentric inversion or by an activation of an evolutionary new centromere. As in the case of MAG2/8, additional investigations with subchromosomal paints or BAC probes are needed in order to find the ancestral position of the centromere as well as to identify the rearrangements that caused centromere repositioning.
It is worth mentioning that A. amphibius (Fig. 7) and D. torquatus (Fig. 2) both have the MAG1/7 association in their karyotypes (MAG7/1/17 in AAM 1 and MAG1/7 in DTO 4). Based on the localization of other sets of probes, this association either represents a conserved block common to these two species or the result of convergence. It is also noteworthy that ENCs at A. oeconomus 11, D. torquatus 10, and Ellobius talpinus 13 were revealed previously using microdissected probes obtained for some mouse chromosomes (Trifonov et al. 2010). This indicates that the emergence of ENCs is not rare in Arvicolinae. Novel ENCs might be revealed by a detailed comparison of other arvicoline species.
Insight into the ancestral karyotype of Arvicolinae
To a large extent, the systematic relationships between Arvicolinae species remain controversial. Phylogenetic trees based on mitochondrial sequences display a different branching order compared to trees built using nuclear sequences, or a combination of mitochondrial and nuclear sequences (Cook et al. 2004; Galewski et al. 2006; Abramson et al. 2009a). However, Galewski et al. (2006) and Abramson et al. (2009b) agree on the phylogenetic relationships reported here: the genus Dicrostonyx separated first from the root of the Arvicolinae tree, followed by the genus Myodes, with genera Arvicola and Microtus being on the most recent branches (Fig. 11). The tribe Ellobiusini, which includes the genus Ellobius, was involved in molecular analysis only in one previous publication (Abramson et al. 2009b). The use of multiple sets of painting probes allowed us to scrutinize some of the chromosomal syntenic associations detected earlier and to provide further insight into the karyotypic relationships between Arvicolinae species.
Our earlier analysis of shared syntenic associations in arvicoline karyotypes allowed us the opportunity to construct ancestral karyotypes for the genera Microtus (Lemskaya et al. 2010) and Ellobius (Romanenko et al. 2007). Expanding the list of species studied and the inclusion of species from “early” branches enable us to build ancestral karyotypes for the taxa below subfamily level. Our comparative chromosome maps constructed for M. agrestis and M. auratus together with the painting data obtained for Arvicolinae and Cricetinae provide an opportunity to reconstruct the putative ancestral Arvicolinae karyotype (AAK).
On the whole, the AAK structure is similar to AMiK (Lemskaya et al. 2010). An important question concerns the number of MAG1 segments. The number of other segments homologous to MAG is similar in AMiK and AAK. There are two possible variants as follows: (a) there were three ancestral MAG1 segments of the AAK conserved in the genomes of D. torquatus (in DTO 4, 5 & 8), Ellobius talpinus (in ETA 5, 9, and 18), in most of the hamster species and mouse; (b) two MAG1 segments presented in the ancestral karyotype (and conserved in other arvicolines so far studied) with one of them subsequently disrupted in some lineages. However, in karyotypes of A. amphibius, M. rutilus, and A. oeconomus fusion MAG 1 peric/1 int + 17 dist (= DTO4/5) which reduce the number of MAG1 fragments in the species karyotypes might have appeared later in the evolution of arvicolines, probably after the separation of the branch leading to D. torquatus. As D. torquatus presents a basal branch of the Arvicolinae tree according to molecular data (Galewski et al. 2006; Abramson et al. 2009b), we postulate here a diploid chromosome number of 2n = 56 and three segments homologous to MAG1 in the AAK (Fig. 12).
The putative ancestral Arvicolinae karyotype (AAK). Single chromosomes of M. agrestis are represented by one element in AAK are shown in blue. The chromosomes represented by two elements are marked by pairs in various colors. Numbers of M. agrestis chromosomes are shown on the right, and their correspondence to M. auratus chromosomes is presented on the left
The question about the MAG1/17 composition in AAK remains open. At this stage of research it seems more likely that MAG1 int/17 (= DTO5) was present in the AAK. The synteny MAG1 prox + int/17(= DTO4/5) appeared later in the evolution of arvicolines, probably after separation of the branch leading to D. torquatus (Fig. 11).
In general, the karyotype evolution in Arvicolinae indicates the preservation of some large conserved segments and many intrachromosomal rearrangements in some conserved segments. The most salient case is the association MAG2/8. This segment is homologous to MAG2/8 prox = DTO1 (Fig. 10) and is undoubtedly present in AAK but its ancestral composition could not be defined without data from different arvicoline species using region-specific probes.
In view of these findings, we postulate that the AAK structure is very similar to AMiK (Fig. 12, Lemskaya et al. 2010). The difference in the diploid chromosome number between AAK and AMiK is caused by a break in the MAG1 segment. Moreover, the synteny MAG13 = DTO12 = MAU3/11 in AAK is different from that in AMiK. It seems more likely that the inversion in the segment MAG13 = DTO12 = MAU11/3/11 appeared later in Arvicolinae evolution or might have arisen independently in separate branches. In AAK, the synteny MAG13 corresponds to DTO12 = MAU3/11. The compositions of syntenies homologous to MAG1/17, 2/8, and 3 remain unclear.
In summary, the extension of comparative chromosome painting to species from the early branching of Arvicolinae here provides a strong support for the structure of the deduced ancestral karyotypes. However, cytogenetic signature markers for the genera have not yet been found. We believe that future high-resolution mapping will uncover such signature rearrangements among undetected cryptic intrachromosomal exchanges. In-depth analysis using a combination of molecular, cytogenetic, and sequence data is needed to demonstrate whether intrachromosomal exchanges indeed played an important role in the rapid and intense speciation of Arvicolinae. This will open a new perspective in comparative genomic and chromosomal studies.
Abbreviations
- AAM:
-
Arvicola amphibius
- AOE:
-
Alexandromys oeconomus (here and further in the text, the species nomenclature is used in accordance with the latest checklist “The mammals of Russia: a taxonomic and geographic reference” (Pavlinov and Lissovsky 2012))
- MRU:
-
Myodes rutilus
- dist:
-
Distal
- DTO:
-
Dicrostonyx torquatus
- ENC:
-
Evolutionary new centromeres
- FISH:
-
Fluorescence in situ hybridization
- GTG-banding:
-
G-banding by trypsin using Giemsa
- int:
-
Interstitial
- ITS:
-
Interstitial telomeric sequences
- MAG:
-
Microtus agrestis
- MAU:
-
Mesocricetus auratus
- prox:
-
Proximal
References
Abramson NI, Lebedev VS, Bannikova AA, Tesakov AS (2009a) Radiation events in the subfamily Arvicolinae (Rodentia): evidence from nuclear genes. Dokl Biol Sci 428:458–461
Abramson NI, Lebedev VS, Tesakov AS, Bannikova AA (2009b) Supraspecies relationships in the subfamily (Rodentia, Cricetidae, Arvicolinae): unexpexted result of nuclear genes analysis. Mol Biol (Mosk) 43:897–909
Abramson NI, Golenishchev FN, Kostygov AYu, Tesakov AS (2011) Taxonomic interpretation of molecular-genetic cladogram for voles of the tribe Mocrotini (Arvicolinae, Rodentia) inferred from nuclear genes. In: Theriofauna of Russia and adjacement regions, 9th Congress of Theriological Society. KMK Sci. Press, Moscow, p. 7
Acosta MJ, Marchal JA, Fernandez-Espartero C et al (2010) Characterization of the satellite DNA Msat-160 from species of Terricola (Microtus) and Arvicola (Rodentia, Arvicolinae). Genetica 138:1085–1098
Bakloushinskaya IY, Matveevsky SN, Romanenko SA et al (2012) A comparative analysis of the mole vole sibling species Ellobius tancrei and E. talpinus (Cricetidae, Rodentia) through chromosome painting and examination of synaptonemal complex structures in hybrids. Cytogenet Genome Res 136:199–207
Bannikova AA, Lebedev VS, Lissovsky AA et al (2010) Molecular phylogeny and evolution of the Asian lineage of vole genus Microtus (Rodentia: Arvicolinae) inferred from mitochondrial cytochrome b sequence. Biol J Linn Soc 99:595–613
Borodin PM, Sablina OV, Rodionova MI (1995) Pattern of X-Y chromosome pairing in microtine rodents. Hereditas 123:17–23
Carleton MD, Musser GG (2005) Subfamily Arvicolinae. In: Wilson E, Reeder D-AM (eds) Mammal species of the world: a taxonomic and geographic reference. Johns Hopkins University Press, Baltimore, pp. 956--1039
Chaline J, Graf JD (1988) Phylogeny of the Arvicolidae (Rodentia)—biochemical and paleontological evidence. J Mammal 69:22–33
Chernyavsky FB, Kozlovsky AI (1980) Species status and history of the Arctic lemming (Dicrostonyx, Rodentia) of Wrangel Island. Zool Zh (in Russian) 59:266–273
Conroy C, Cook JA (1999) MtDNA evidence for repeated pulses of speciation within arvicoline and murid rodents. J Mamm Evol 6:221–245
Cook JA, Runck AM, Conroy CJ (2004) Historical biogeography at the crossroads of the northern continents: molecular phylogenetics of red-backed voles (Rodentia: Arvicolinae). Mol Phylogenet Evol 30:767–777
de la Fuente R, Sanchez A, Marchal JA et al (2012) A synaptonemal complex-derived mechanism for meiotic segregation precedes the evolutionary loss of homology between sex chromosomes in arvicolid mammals. Chromosoma 121:433–446
DeWoody JA, Chesser RK, Baker RJ (1999) A translocated mitochondrial cytochrome b pseudogene in voles (Rodentia: Microtus). J Mol Evol 48:380–382
Fredga K (1983) Aberrant sex chromosome mechanisms in mammals. Evol asp Differ 23(Suppl):S23–30
Fredga K, Fedorov V, Jarrell G, Jonsson L (1999) Genetic diversity in Arctic lemmings. Ambio 28:261--269
Galewski T, Tilak MK, Sanchez S et al (2006) The evolutionary radiation of Arvicolinae rodents (voles and lemmings): relative contribution of nuclear and mitochondrial DNA phylogenies. BMC Evol Biol 6:80
Gileva EA (1980) Chromosomal diversity and an aberrant genetic system of sex determination in the arctic lemming, Dicrostonyx-torquatus pallas (1779). Genetica 52–3:99–103
Gileva EA (1983) A contrasted pattern of chromosome evolution in 2 genera of lemmings, Lemmus and Dicrostonyx (Mammalia, Rodentia). Genetica 60:173–179
Gileva EA (2004) The B chromosome system in the varying lemming Dicrostonyx torquatus pall., 1779 from natural and laboratory populations. Russ J Genet 40:1399–1406
Gileva EA, Chebotar NA (1979) Fertile XO males and females in the varying lemming, Dicrostonyx torquatus pall. 1779. Heredity 42:67–77
Graphodatsky AS, Sablina OV, Meyer MN et al (2000) Comparative cytogenetics of hamsters of the genus Calomyscus. Cytogenet Cell Genet 88:296–304
Ijdo JW, Wells RA, Baldini A, Reeders ST (1991) Improved telomere detection using a telomere repeat probe (TTAGGG) n generated by PCR. Nucleic Acids Res 19:4780
Jaarola M, Martinkova N, Gunduz I et al (2004) Molecular phylogeny of the speciose vole genus Microtus (Arvicolinae, Rodentia) inferred from mitochondrial DNA sequences. Mol Phylogenet Evol 33:647–663
Lemskaya NA, Romanenko SA, Golenishchev FN et al (2010) Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). III. Karyotype relationships of ten Microtus species. Chromosome Res 18:459–471
Li T, Wang J, Su W, Nie W, Yang F (2006) Karyotypic evolution of the family Sciuridae: inferences from the genome organizations of ground squirrels. Cytogenet Genome Res 112:270–276
Maden BE, Dent CL, Farrell TE et al (1987) Clones of human ribosomal DNA containing the complete 18 S-rRNA and 28 S-rRNA genes. Characterization, a detailed map of the human ribosomal transcription unit and diversity among clones. Biochem J 246:519–527
Mazurok NA, Rubtsova NV, Isaenko AA et al (2001) Comparative chromosome and mitochondrial DNA analyses and phylogenetic relationships within common voles (Microtus, Arvicolidae). Chromosome Res 9:107–120
Modi WS (1987a) C-banding analyses and the evolution of heterochromatin among arvicolid rodents. J Mammal 68:704–714
Modi WS (1987b) Phylogenetic analyses of chromosomal banding-patterns among the Nearctic Arvicolidae (Mammalia, Rodentia). Syst Zool 36:109–136
Orlov VN, Bulatova NSh (1983) Comparative cytogenetics and karyosystematic of mammals. Nauka, Moscow (In russian)
Pavlinov IYa, Lissovsky AA (2012) The mammals of Russia: a taxonomic and geographic reference. In: Kalyakin MV (ed) Archives of zoological museum of Moscow state university. KMK Scientific Press Ltd, Moscow, pp. 1--604
Repenning CA (1990) Of mice and ice in the Late Pliocene of North-America. Arctic 43:314--323
Romanenko SA, Volobouev V (2012) Non-sciuromorph rodent karyotypes in evolution. Cytogenet Genome Res 137:233–245
Romanenko SA, Perelman PL, Serdukova NA et al (2006) Reciprocal chromosome painting between three laboratory rodent species. Mamm Genome 17:1183–1192
Romanenko SA, Sitnikova NA, Serdukova NA et al (2007) Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). II. The genome homology of two mole voles (genus Ellobius), the field vole and golden hamster revealed by comparative chromosome painting. Chromosome Res 15:891–897
Romanenko SA, Perelman PL, Trifonov VA, Graphodatsky AS (2012) Chromosomal evolution in Rodentia. Heredity (Edinb) 108:4–16
Seabright M (1971) A rapid banding technique for human chromosomes. Lancet 11:971–972
Sitnikova NA, Romanenko SA, O'Brien PC et al (2007) Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). I. The genome homology of tundra vole, field vole, mouse and golden hamster revealed by comparative chromosome painting. Chromosome Res 15:447–456
Telenius H, Pelmear AH, Tunnacliffe A et al (1992) Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosom Cancer 4:257–263
Triant DA, Dewoody JA (2006) Accelerated molecular evolution in Microtus (Rodentia) as assessed via complete mitochondrial genome sequences. Genetica 128:95–108
Trifonov VA, Kosyakova N, Romanenko SA et al (2010) New insights into the karyotypic evolution in muroid rodents revealed by multicolor banding applying murine probes. Chromosom Res 18:265–275
Weimer J, Kiechle M, Arnold N (2000) FISH-microdissection (FISH-MD) analysis of complex chromosome rearrangements. Cytogenet Cell Genet 88:114–118
Yang F, Carter NP, Shi L, Ferguson-Smith MA (1995) A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma 103:642–652
Yang F, O'Brien PC, Milne BS et al (1999) A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 62:189–202
Yannic G, Burri R, Malikov VG, Vogel P (2012) Systematics of snow voles (Chionomys, Arvicolinae) revisited. Mol Phylogenet Evol 62:806–815
Acknowledgments
This study was funded in part by the MCB and SB RAS Programs, research grants of Russian Fund for Basic Research (No. 11-04-00673, No. 14-04-00451 (SAR); No. 14-04-31555 (NAL); No. 15-29-02384, No. 15-04-00962 (ASG)) and ZIN RAS (project No. 01201351185).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All institutional and national guidelines for the care and use of laboratory animals were followed.
Conflict of interest
The authors declare that they have no competing of interests.
Additional information
Responsible Editor: Irina Solovei, PhD
Rights and permissions
About this article
Cite this article
Romanenko, S.A., Lemskaya, N.A., Trifonov, V.A. et al. Genome-wide comparative chromosome maps of Arvicola amphibius, Dicrostonyx torquatus, and Myodes rutilus . Chromosome Res 24, 145–159 (2016). https://doi.org/10.1007/s10577-015-9504-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-015-9504-6