Abstract
The laboratory mouse (Mus musculus, 2n = 40), the Chinese hamster (Cricetulus griseus, 2n = 22), and the golden (Syrian) hamster (Mesocricetus auratus, 2n = 44) are common laboratory animals, extensively used in biomedical research. In contrast with the mouse genome, which was sequenced and well characterized, the hamster species has been set aside. We constructed a chromosome paint set for the golden hamster, which for the first time allowed us to perform multidirectional chromosome painting between the golden hamster and the mouse and between the two species of hamster. From these data we constructed a detailed comparative chromosome map of the laboratory mouse and the two hamster species. The golden hamster painting probes revealed 25 autosomal segments in the Chinese hamster and 43 in the mouse. Using the Chinese hamster probes, 23 conserved segments were found in the golden hamster karyotype. The mouse probes revealed 42 conserved autosomal segments in the golden hamster karyotype. The two largest chromosomes of the Chinese hamster (1 and 2) are homologous to seven and five chromosomes of the golden hamster, respectively. The golden hamster karyotype can be transformed into the Chinese hamster karyotype by 15 fusions and 3 fissions. Previous reconstructions of the ancestral murid karyotype proposed diploid numbers from 2n = 52 to 2n = 54. By integrating the new multidirectional chromosome painting data presented here with previous comparative genomics data, we can propose that syntenies to mouse Chrs 6 and 16 were both present and to hypothesize a diploid number of 2n = 48 for the ancestral Murinae/Cricetinae karyotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myomorph rodents, e.g., rats, mice, voles, hamsters, jerboas, and many allied forms, are a particular group of species characterized by extremely high rates of chromosome evolution and remarkable chromosome polymorphism (wide variation in diploid numbers, heterochromatin quantity, and possession of supernumerary B chromosomes) (Graphodatsky 1989; Murphy et al. 2001). This has made myomorph rodents difficult to study using comparative chromosome analysis (Scherthan et al. 1994). Cross-species chromosome painting within the myomoph group has been done only for some species of the Muridae family: Mus musculus and Rattus norvegicus (Guilly et al. 1999, 2001; Stanyon et al. 1999), R. rattus rattus and R. rattus frugivorous (Cavagna et al. 2002), M. musculus and C. griseus (Yang et al. 2000), M. musculus and Apodemus sylvaticus (Stanyon et al. 2004), M. musculus and Mus platythrix (Matsubara et al. 2003), M. musculus and Otomys irroratus (Engelbrecht et al. 2006), M. musculus and Rhabdomys pumilio (Rambau and Robinson 2003), and M. musculus and seven species of Apodemus (Matsubara et al. 2004). Homologies between some chromosomes of M. musculus and Peromyscus maniculatus were partly defined (Dawson et al. 1999).
In earlier comparative studies of chromosome G-banding patterns, some homologous elements in the mouse and the Chinese hamster karyotypes were found (Gamperl et al. 1978; Graphodatsky 1989). However, no genome-wide comparison of these two karyotypes was possible until reciprocal painting between rodent species from Cricetinae (Chinese hamster) and Murinae (mouse) subfamilies confirmed intensive chromosome rearrangements during their karyotype evolution (Yang et al. 2000). No data have been reported so far on chromosome painting of the golden hamster.
In this article we present a chromosome comparison of the laboratory mouse (Mus musculus, 2n = 40), the golden hamster (Mesocricetus auratus, 2n = 44), and the Chinese hamster (Cricetulus griseus, 2n = 22) using three sets of painting probes. Here golden hamster painting probes were used for the first time to compare karyotypes of these rodents in detail. We performed the reciprocal painting between the Chinese hamster and golden hamster and between the mouse and golden hamster karyotypes.
Materials and methods
Animals
Chromosome suspensions were obtained from a fibroblast cell line in the Laboratory of Human and Animals Cytogenetics, the Institute of Cytology and Genetics, Russia, and in the Museum National d’Histoire Naturelle, Origine, Structure et Evolution de la Biodiversite, Paris, France.
Cell culture and metaphase preparation
Fibroblast cultures were established from male specimens of mouse, Chinese hamster, and golden hamster. Cells were cultivated in alpha-DMEM supplemented with 10% fetal bovine serum. Metaphase chromosome spreads were prepared from primary fibroblast cultures as described previously (Yang et al. 1999, Graphodatsky et al. 2000, 2001).
Nomenclature of golden hamster chromosomes
The golden hamster (M. auratus) karyotype was reported earlier but no standard nomenclature of chromosomes has been proposed (Popescu and DiPaolo 1972; Pavia et al. 1977; Li et al. 1982; http://www.bionet.nsc.ru). The diploid chromosome number (2n = 44) and fundamental number found in this study were the same as those reported previously. The chromosomes are numbered according to Radjabli et al. (2006).
Painting probes
We used painting probes derived from the mouse and the Chinese and golden hamsters to perform cross-species chromosome painting. The sets of mouse (Yang et al. 2000; Stanyon et al. 2004) and Chinese hamster (Yang et al. 2000) paints reported previously were used in this study. Briefly, this set of Chinese hamster painting probes consisted of nine probes, each representing one chromosome, and one probe containing both autosomes 9 and 10. Both sets of laboratory mouse painting probes consisted of 20 probes and represented all individual autosomes and the X chromosome. Hybridizing the probes onto G-banded chromosomes of the same species identified the content of each paint.
The golden hamster paints were generated in the Key Laboratory of Cellular and Molecular Evolution, Chinese Academy of Sciences, by degenerative oligonucleotide priming (DOP)-PCR amplification of flow-sorted chromosomes (Telenius et al. 1992; Yang et al. 1995) with the exception of the painting probes for Chromosomes 15, 16, and 21 of golden hamster, which were obtained by microdissection as described (Trifonov et al. 2002).
Fish
G-banding was performed before fluorescence in situ hybridization (FISH), using the standard procedure (Seabright 1971). Metaphases were photographed, slides destained in methanol, and fixed with 1% formaldehyde. FISH was performed using a standard protocol (Yang et al. 1999, Graphodatsky et al. 2000). In short, for each hybridization experiment we used 0.2 μg biotinylated DOP-PCR product and 30 μg Cot-5 DNA of M. auratus in 15 μl of hybridization buffer (10% dextran sulfate, 50% formamide, 2×SSC). Paints were denatured at 96°C for 3 min and reannealed for 30 min at 42°C. Probes were hybridized overnight at 42°C. Posthybridization washes included 40% formamide, 2×SSC 3 times, 2×SSC 3 times (5 min each), followed by 30-min incubation in 3% dry milk in 4×SSC/0.1% Triton X-100. All washes were performed at 42°C. Hybridization signals were detected with avidin-FITC without further amplification. Chromosomes were counterstained with DAPI.
Image capture
Images were captured using In Situ Imaging System (ISIS) software (Metasystems) with a Paco CCD camera mounted on an Aristoplan (Leitz) microscope. The microscope was equipped with two filter sets for FITC and DAPI, respectively. Fluorescence signals were captured separately as 8-bit black-and-white images through appropriate excitation filters, normalized, and merged to a 24-bit color image. Hybridization signals were assigned to specific chromosome regions defined by G-banding patterns previously photographed and captured by the CCD camera. All image processing was performed using Paint Shop Pro (Jasc Software).
Results
Golden hamster flow karyotype
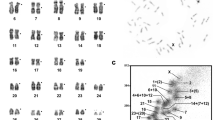
The set of golden hamster (M. auratus, 2n = 44) chromosomes was separated into 17 regions (Fig. 1). As the karyotype of the golden hamster contains many metacentric chromosomes with similar size and base ratio, it was difficult to resolve some individual chromosomes by sorting. The content of each peak was identified by hybridizing the probes onto G-banded chromosomes of the golden hamster. Eleven peaks each contained a particular chromosome (1, 2, 3, 5, 7, 8, 9, 10, 17, 18, and 20); four peaks provided hybridization signals on two chromosomes (X and Y, 4 and 6, 12 and 13, 18 and 19), while two probes highlighted three and four chromosomes (11, 14, 15 and 11, 14–16). To refine the resolution of analysis, microdissection of some golden hamster chromosomes (15, 16, and 21) was performed. Thus, 20 painting probes, containing all 19 golden hamster autosomes and the X chromosome, were used in this study: 1, 2, 3, 4+6, 5, 7, 8, 9, 10, 11+14+15, 11+14+15+16, 12 +13, 15, 16, 17, 18, 18+19, 20, 21, and X.
Reciprocal chromosome painting of Chinese and golden hamsters
Examples of FISH are shown in Fig. 2a, b. The 20 painting probes of the golden hamster revealed 25 conserved autosomal segments in the genome of the Chinese hamster (Fig. 3). Almost all probes (Chrs 1–4, 6–8, 10, 12, 13, 15–21, and X) delineated one region on the Chinese hamster chromosomes. Other golden hamster probes (Chrs 5, 9, 11, 14) each delineated two regions in the C. griseus karyotype. The reciprocal hybridization of nine Chinese hamster painting probes produced 23 signals on golden hamster autosomes (Fig. 4). Five Chinese hamster chromosome probes (6–9 and X) painted one region on the golden hamster karyotype, three probes (Chrs 3–5) each delineated two regions on golden hamster autosomes, and two probes (Chrs 1 and 2) painted seven and five regions, respectively.
Examples of fluorescent in situ hybridization: (a) localization of painting probe of Chr 5 of M. auratus on C. griseus chromosomes; (b) localization of painting probe of microdissected Chr 16 of M. auratus on C. griseus chromosomes; (c) localization of M. auratus Chr 2 on M. musculus; (d) localization of M. musculus Chr 1 on M. auratus chromosomes.
Comparative chromosome map of Chinese hamster (CGR, banded chromosomes at the left) and golden hamster (MAU, black line) established by reciprocal hybridization of golden and Chinese hamster chromosome-specific painting probes. At the right is presented correspondence to mouse chromosomes (MMU, black line) established by Yang et al. (2000) and refined by reciprocal painting of mouse and golden hamster chromosome-specific painting probes.
Even though there are twice as many chromosomes in the golden hamster (2n = 44) as in the Chinese hamster (2n = 22), homologous regions defined by chromosome painting showed good correspondences in their banding patterns, suggesting that there were limited intrachromosomal rearrangements. However, for some segments on Chrs 1 and 2 of the Chinese hamster, it was hard to show homology of the banding pattern because of the small size of fragments.
The two largest chromosomes of the Chinese hamster (Chrs 1 and 2) are homologous to seven and five golden hamster chromosomes, respectively (Fig. 3). Chromosomes 5 and 14 of the golden hamster both contain the association 1 and 2 of C. griseus chromosome segments (Fig. 4). The golden hamster karyotype can be formed from the Chinese hamster karyotype by 13 fissions and 2 fusions, whereas 15 fusions and 3 fissions are needed to transform the golden hamster karyotype into the Chinese hamster karyotype.
Reciprocal chromosome painting of mouse and golden hamster
The 20 painting probes of golden hamster revealed 43 conserved autosomal segments in the mouse genome (Fig. 5). The whole set of mouse chromosome-specific probes revealed 42 conserved autosomal segments in the golden hamster karyotype (Fig. 4). Examples of FISH are shown in Fig. 3c, d. Three probes (mouse Chrs 9, 19, and X) each painted one chromosomal region of M. auratus. Thirteen probes (mouse Chrs 2–8, 11–14, 16, and 18) each delineated two regions on the golden hamster karyotype. The mouse probes 1, 10, and 15 painted three regions while probe 17 painted 5 regions in the M. auratus genome.
The correspondence of mouse chromosomes (Mmu, on the left) and homologic segments of C. griseus (CGR, in the middle) by Yang et al. (2000) and M. auratus (MAU, on the right) established by the reciprocal painting of mouse and golden hamster chromsomes.
Reciprocal hybridization of mouse and golden hamster chromosome paints revealed the following associations of M. auratus chromosomal segments in M. musculus: 5pa/5qa/8a, 1/7a, 17/19a, 3a/13, 4a/16a, 6/9a, 2a/21, 7b/9b, 14a/5pb/19b, 10a/14b, 10b/18, 7c/11a, 15/16b, 5qb/20/11b, 4b/10c, 14c/10d/5pc/8b/10e/5qc, 3b/11c (Fig. 5). Twelve mouse chromosome associations are present in the golden hamster karyotype (Fig. 4). Mouse Chromosomes 1 and 17 painted disjunct regions on Chromosome 5 of the golden hamster with signals on both the p and q arms. The most probable explanation for this pattern is that an inversion occurred in the golden hamster karyotype.
Discussion
This article presents the first report on reciprocal painting between two species from the Cricetinae, the golden hamster and the Chinese hamster. We also used multidirectional chromosome painting to provide a more accurate and full assessment of chromosomal rearrangements and breakpoints between three laboratory rodent species (mouse, Chinese hamster, and golden hamster). These data provide new information for the reconstruction of the ancestral Murinae/Cricetinae karyotype, which may well have implications for the content of the ancestral muroidea genome.
Murinae and Cricetinae karyotypic relationships
Earlier chromosomal comparison of mouse (Murinae) and Chinese hamster (Cricetinae) by reciprocal chromosome painting was carried out and a comparative chromosome map for these two species was built (Yang et al. 2000). The use of three different sets of painting probes along with published data on these previous reciprocal data permitted us to make a more detailed chromosome comparison of M. auratus, C. griseus, and M. musculus and find additional intrachromosomal rearrangements distinguishing the hamsters and mouse karyotypes.
The comparative chromosome map for these three rodent species defines regions of homology between the karyotypes (Fig. 6). It is interesting that although Chromosome 7 of C. griseus seems to have almost the same banding pattern as Chromosome 10 of M. auratus, the distribution of mouse chromosome segments on these homologous chromosomes varies. This indicates possible inversion events (at least three rearrangements are necessary to convert the segment order) (Fig. 6). Clearly, apparently conserved or identical banding patterns permit hypotheses of conserved gene order, which need to be tested at the molecular level. In the case of fusion 13/15 of mouse chromosomes on C. griseus Chromosome 2, we suggest that an inversion occurred in the Chinese hamster.
G-banding comparisons of chromosomes between M. musculus (Mmu), C. griseus (Cgr), and M. auratus (Mau) as identified by FISH. Bold numbers mark C. griseus chromosomes. Chromosomes of M. musculus and M. auratus are indicated by italic and roman fonts, respectively. The correspondence of C. griseus Chrs 3–6, 8–10, and X (CGR, in the middle) and homologic segments of M. musculus (on the left) and M. auratus (on the right) are shown by lines. The correspondence of the most complex chromosomes of C. griseus (1, 2, and 7) with M. musculus and M. auratus chromosomes is shown separately within a framework.
Chromosome 9 of the golden hamster revealed two homologous segments on Chinese hamster Chr 1. It is possible that the segments were derived by an inversion. However, because these segments are present in different mouse chromosomes (MMU6 and MMU8), one could suggest that they were present as two chromosomes in the common ancestor of hamsters and mouse. M. auratus Chr 11 is present on two regions of C. griseus Chr 2 and gives three signals in the mouse genome (MMU13, 15, and 18). The pattern of the probe distribution may indicate that MAU11 was present as two chromosomes in an ancestor Eumuroida karyotype (corresponding to MMU13/15 and MMU18).
Implication for the ancestral Eumuroida karyo- type
There are 14 rodent species studied up to now using mouse painting probes (Guilly et al. 1999, 2001; Stanyon et al. 1999, 2004; Yang et al. 2000; Cavagna et al. 2002; Rambau and Robinson 2003; Matsubara et al. 2003; Engelbrecht et al. 2006).
All reported species belong to two subfamilies the Murinae (7 Apodemus species, Otomys, Rhabdomys, 2 Rattus species,2 Mus species) and Cricetinae (Cricetulus) within the large superfamily Eumuroida (Steppan et al. 2004). These data allow speculation about the course of chromosome evolution in this group. The data may also have implications for some previous hypothesis concerning the ancestral Eumuroida karyotype (AEK). However, it should be noted that the current sample of taxa is still extremely thin for one of the largest mammalian clades. All conclusions can be considered working hypotheses which can help clarify future research directions. The presence of the same combination of chromosome segments in different species may indicate the ancestral origin of the syntenic group. Yang et al. (2000) suggested earlier that associations of mouse chromosomal segments 7/19 (AEK1), 13/15 (AEK14), 12/17 (AEK22), 17/1/17 (AEK9), 5/11 (AEK8), 2/13 (AEK16), and 11/17 may represent ancestral syntenies for rodents with mouse Chrs 3 (AEK4), 4 (AEK5), 9 (AEK7), 18 (AEK12), and 19 being presented as single chromosomes.
According to another hypothesis, the ancestral karyotype of eumuroid rodents had a diploid number of 2n = 54 and contained the following chromosomes (composed from homologous mouse chromosome segments): MMU 1(AEK13), 1/17 (AEK9), 2 (AEK3), 2/13 (AEK16), 3 (AEK4), 4 (AEK5), 5 (AEK 19), 5/11 (AEK 8), 6a, 6b, 7/19 (AEK1), 8a (AEK17), 8b (AEK18), 9 (AEK7), 10a (AEK23), 10b/17b (AEK21), 10c/17c (AEK20), 11, 11/16, 12a (AEK11), 12b/17d (AEK22), 13b/15a (AEK14), 14 (AEK10), 15b (AEK15), 16b, 18 (AEK12), X, and Y (Stanyon et al. 2004). The hypothesis was based on the comparative analysis of M. musculus painting data on five Murinae and one Criceninae species. Our ancestral karyotype differs in the composition of two chromosomes: AEK 2 (16/11b) and AEK 6 (Fig. 7).
Reconstructed ancestral Eumuroida karyotype. The homologies of individual conserved segments to mouse and golden hamster chromosomes are shown to the left and to the right of the ideograms, respectively. Beside each chromosome of the ancestor is shown its possible homology with conforming fragments of human chromosomes.
A more precise structure of the ancestral Eumuroida karyotype can be defined if a species from another order is used as an outgroup. In the present study the use of the human karyotype seemed convenient. The high level of fragmentation and repatterning of the mouse genome probably accounts for the limited success of chromosome reciprocal painting between human and mouse (Ferguson-Smith 1997). However, gene mapping and sequencing data are now available to compare the highly rearranged genomes of muroid rodents with the human genome. Using the data in Murphy et al. (2005), more then 60 mouse chromosome associations were defined for the human karyotype. Genome browsers provide even more detailed comparative maps between the human and mouse genomes and probably a higher number of associations could be described, far beyond the resolution provided by cytogenetics. For example, see mouse–human orthology map with the 113 human homologous segments mapped onto 20 Mus musculus chromosomes (http://www.informatics.jax.org/reports/homologymap/mouse_human.shtml).
The addition of a new hamster species (Mesocricetus auratus, Cricetinae) together with human and mouse genome comparisons and some of our unpublished results allowed us to test the previously suggested ancestral karyotypes (Yang et al. 2000; Stanyon et al. 2004; Engelbrecht et al. 2006) and to propose for discussion the new hypothetical Eumuroida ancestor karyotype with 2n = 48 (Fig. 7). As the basis for construction of this karyotype, analysis of the presence or absence of chromosomal associations has served in karyotypes of the Eumuroida studied until now. In addition, we have added for this analysis unpublished data on Peromyscus, Calomyscus, and Microtus species (Table 1).
We propose that the ancestral Eumuroida karyotype had 2n = 48 and contained the following murine chromosomal segments and associations: 1a/17a (MAU5a), 1b (MAU8?), 2a (MAU1), 2b/13a (MAU7a), 3 (MAU17/19a), 4 (MAU3a/13), 5a/11a (MAU16a?/14a), 5b (MAU4a), 6 (MAU6/9a), 7/19 (MAU21/2), 8a (MAU9b), 8b (MAU7b), 9 (MAU12), 10a (MAU19b), 10b/17b (MAU5b), 10c/17c (MAU14b), 11b/16 (MAU4b/10a), 12a (MAU18), 12b/17d (MAU10b), 13b/15a (MAU11a), 14 (MAU15/16b?), 15b (MAU5c/20), 18 (MAU3b/11b), X, Y. Future data from more species will contribute to more reliable reconstruction of the ancestral Eumuroida karyotype, provide insights into phylogeny, and answer questions of the mode of karyotypic changes in rodents.
References
Cavagna P, Stone G, Stanyon R (2002) Black rat (Rattus rattus) genomic variability characterized by chromosome painting. Mamm Genome 13, 157–163
Dawson WD, Young SR, Wang Z, Liu LW, Greenbaum IF, et al. (1999) Mus and Peromyscus chromosome homology established by FISH with three mouse paint probes. Mamm Genome 10, 730–733
Engelbrecht A, Dobigny G, Robinson TJ (2006) Further insights into the ancestral murine karyotype: the contribution of the Otomys-Mus comparison using chromosome painting. Cytogenet Genome Res 112, 126–130
Ferguson-Smith MA (1997) Genetic analysis by chromosome sorting and painting: phylogenetic and diagnostic applications. Eur J Human Genet 5, 253–265
Gamperl R, Vistorin G, Rosenkranz W (1978) Comparison of chromosome banding patterns in five members of Cricetinae with comments on possible relationships. Caryologia 31, 343–353
Graphodatsky AS (1989) Conserved and variable elements of mammalian chromosomes. In Cytogenetics of Animals, Halnan C, (ed.) (Oxon: CAB International Press), pp 95–123
Graphodatsky AS, Sablina OV, Meyer MN, Malikov VG, Isakova EA, et al. (2000) Comparative cytogenetics of hamsters of the genus Calomyscus. Cytogenet Cell Genet 88, 296–304
Graphodatsky AS, Yang F, O’Brien PC, Perelman P, Milne BS., et al. (2001) Phylogenetic implications of the 38 putative ancestral chromosome segments for four canid species. Cytogenet Cell Genet 92, 243–247
Guilly M-N, Fouchet P, de Chamisso P, Schmitz A, Dutrillaux B (1999) Comparative karyotype of rat and mouse using bidirectional chromosome painting. Chromosome Res 7, 213–221
Guilly M-N, Dano L, de Chamisso P, Fouchet P, Dutrillaux B, et al. (2001) Comparative karyotype using bidirectional chromosome painting: how and why? Meth Cell Sci 23, 163–170
Li S, Pathak S, Hsu TC (1982) High resolution G-banding patterns of Syrian hamster chromosomes. Cytogenet Cell Genet 33, 295–302
Matsubara K, Nishida-Umehara C, Kuroiwa A, Tsuchiya K, Matsuda Y (2003) Identification of chromosome rearrangements between the laboratory mouse (Mus musculus) and the Indian spiny mouse (Mus platythrix) by comparative FISH analysis. Chromosome Res 11, 57–64
Matsubara K, Nishida-Umehara C, Tsuchiya K, Nukaya D, Matsuda Y (2004) Karyotypic evolution of Apodemus (Muridae, Rodentia) inferred from comparative FISH analyses. Chromosome Res 12, 383–395
Murphy WJ, Stanyon R, O’Brien SJ (2001) Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol 2(6), 1–8
Murphy WJ, Larkin DM, Evert-van der Wind A, Bourque G, Tesler G, et al. (2005) Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309, 613–617
Pavia RA, Smith LW, Goldenberg DM (1977) An analysis of the G-banded chromosomes of the golden hamster. Int J Cancer 20, 460–465
Popescu NC, DiPaolo JA (1972) Identification of Syrian hamster chromosomes by acetic-saline-Giemsa (ASG) and trypsin techniques. Cytogentics 11, 500–507
Radjabli SI, Sablina OV, Graphodatsky AS (2006) Selected karyotypes. In ATLAS of Mammalian Karyotypes, O’Brien SJ, Nash WG, Menninger JC, (eds.) (New York: John Wiley & Sons), pp 305, 329–352
Rambau RV, Robinson TJ (2003) Chromosome painting in the African four-striped mouse Rhabdomys pumilio: detection of possible murid specific contiguous segment combination. Chromosome Res 11, 91–98
Scherthan H, Cremer T, Arnason U, Weier HU, Lima-de-Faria A, et al. (1994) Comparative chromosome painting discloses homologous segments in distantly related mammals. Nat Genet 6(4), 342–347
Seabright M (1971) A rapid banding technique for human chromosomes. Lancet 2, 971–972
Stanyon R, Yang F, Cavagna P, O’Brien PC, Bagga M, et al. (1999) Reciprocal chromosome painting shows that genomic rearrangement between rat and mouse proceeds ten times faster than between humans and cats. Cytogenet Cell Genet 84(3–4), 150–155
Stanyon R, Yang F, Morescalchi AM, Galleni L (2004) Chromosome painting in the long-tailed field mouse provides insights into the ancestral murid karyotype. Cytogenet Genome Res 105, 406–411
Steppan S, Adkins R, Anderson J (2004) Phylogeny and divergence-data estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol 53(4), 533–553
Telenius H, Pelmear AH, Tunnacliffe A, Carter NP, Behmel A, et al. (1992) Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosomes Cancer 4, 226–257
Trifonov VA, Perelman PL, Kawada SI, Iwasa MA, Oda SI, et al. (2002) Complex structure of B-chromosomes in two mammalian species: Apodemus peninsulae (Rodentia) and Nyctereutes procyonoides (Carnivora) revealed by microdissection. Chromosome Res 10(2), 109–116
Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562
Yang F, Carter NP, Shi, Ferguson-Smith MA (1995) A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma 103, 642–652
Yang F, O’Brien PC, Milne BS, Graphodatsky AS, Solanky N, et al. (1999) A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 62, 189–202
Yang F, O’Brien PC, Ferguson-Smith MA (2000) Comparative chromosome map of the laboratory mouse and Chinese hamster defined by reciprocal chromosome painting. Chromosome Res 8, 219–227
Acknowledgments
This study was funded in part by Molecular and Cell Biology (MCB), Russian Fund for Basic Research (RFBR), Biosphere Origin and Evolution (BOE), Dynamics of Genofonds of Plants, Animals and Human (DGPAH) research grants (ASG), an INTAS Grant to ASG and MAF-S, and a Wellcome trust grant to MAF-S. RS and PLP thank Gary Stone for additional flow sorting. RS was previously at NCI-Frederick and was partially supported by a grant from MIUR (Ministero Italiano della Universita’ e della Ricerca), “Mobility of Italian and Foreign Researchers Residing Abroad.”
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Romanenko, S.A., Perelman, P.L., Serdukova, N.A. et al. Reciprocal chromosome painting between three laboratory rodent species. Mamm Genome 17, 1183–1192 (2006). https://doi.org/10.1007/s00335-006-0081-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-006-0081-z