Abstract
Background
Tailoring warfarin use poses a challenge for physicians and pharmacists due to its narrow therapeutic window and substantial inter-individual variability. This study aimed to create an adapted neural-fuzzy inference system (ANFIS) model using preprocessed balance data to improve the predictive accuracy of warfarin maintenance dosing in Chinese patients undergoing heart valve replacement (HVR).
Methods
This retrospective study enrolled patients who underwent HVR between June 1, 2012, and June 1, 2016, from 35 centers in China. The primary outcomes were the mean difference between predicted warfarin dose by ANFIS models and actual dose and the models’ predictive accuracy, including the ideal predicted percentage, the mean absolute error (MAE), and the mean squared error (MSE). The eligible cases were divided into training, internal validation, and external validation groups. We explored input variables by univariate analysis of a general linear model and created two ANFIS models using imbalanced and balanced training sets. We finally compared the primary outcomes between the imbalanced and balanced ANFIS models in both internal and external validation sets. Stratified analyses were conducted across warfarin doses (low, medium, and high doses).
Results
A total of 15,108 patients were included and grouped as follows: 12,086 in the imbalanced training set; 2820 in the balanced training set; 1511 in the internal validation set; and 1511 in the external validation set. Eight variables were explored as predictors related to warfarin maintenance doses, and imbalanced and balanced ANFIS models with multi-fuzzy rules were developed. The results showed a low mean difference between predicted and actual doses (< 0.3 mg/d for each model) and an accurate prediction property in both the imbalanced model (ideal prediction percentage, 74.39–78.16%; MAE, 0.37 mg/daily; MSE, 0.39 mg/daily) and the balanced model (ideal prediction percentage, 73.46–75.31%; MAE, 0.42 mg/daily; MSE, 0.43 mg/daily). Compared to the imbalanced model, the balanced model had a significantly higher prediction accuracy in the low-dose (14.46% vs. 3.01%; P < 0.001) and the high-dose warfarin groups (34.71% vs. 23.14%; P = 0.047). The results from the external validation cohort confirmed this finding.
Conclusions
The ANFIS model can accurately predict the warfarin maintenance dose in patients after HVR. Through data preprocessing, the balanced model contributed to improved prediction ability in the low- and high-dose warfarin groups.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Warfarin, with a clear effectiveness and price advantage, is recommended for the prevention of thrombosis after heart valve replacement (HVR) [1]. However, the need for frequent monitoring, the narrow therapeutic range, dietary restrictions, and multiple drug interactions associated with warfarin have contributed to the insufficiency or excessiveness of anticoagulation, which can lead to thromboembolism and bleeding. Notably, significant individual diversity leads to considerable differences (of up to 20-fold) in the response of patients to warfarin doses [2]. Therefore, a tailored warfarin dose may reduce complications and improve the survival rate of patients with HVR [3].
At present, the warfarin individualized drug prediction model can be divided into multiple linear regression (MLR) and a machine learning algorithm [4]. Specific pharmacogenomic variables, such as cytochrome P450, family 2, subfamily C, polypeptide 9 (CYP2C9) and vitamin K epoxide reductase complex, and subunit 1 (VKORC1), could explain the 55% warfarin dose variation and have been introduced to improve models’ prediction ability [5]. Gage et al. and the International Warfarin Pharmacogenetics Consortium (IWPC) have developed two representatively linear warfarin prediction models based on pharmacogenomic information and clinical factors; these models could explain 43–54% of the variability in the warfarin dose [6, 7]. Currently, there is no warfarin maintenance dose prediction model that is able to achieve an overall prediction accuracy higher than 70% [5, 8, 9].
In China, where medical insurance is limited and there is a lack of primary medical units, extensive gene sequencing for predicting warfarin dose variation will bring unnecessary economic burden, which is contrary to the original cost-efficiency advantage of warfarin. Thus, developing the optimal prediction model for warfarin dosing based on explicable clinical variables poses a challenging task. Unlike the MLR method, machine learning algorithms, such as adapted neural-fuzzy inference system (ANFIS), are data mining methodologies that work based on a combination of fuzzy logic and neural networks. They determine various clustering values in fuzzy sets, predict membership functions during the training process, and calculate the weights by a neural network model. Efficiently, this technique simplifies the problems of complicated characteristics in the process of system modeling and is used in a wide variety of inference problems including those in mechanics and engineering. In previous studies, we established the ANFIS models [10], the artificial neural network (ANN) [11], the back-propagation neural network with genetic algorithm (BPGA) [12], and the back-propagation neural network (BPNN) [13], based on machine learning algorithms, to predict the maintenance dose of warfarin. As a result, we achieved the actual warfarin dose with 59–78% accuracy for patients who underwent AVR. However, we also found poor accuracy for patients in the low- and high-dose groups. Similar results were also observed in other warfarin dose models [14,15,16,17]. One explanation for this might be the class imbalance learning (CIL) problem, which is associated with the size of the data used to train the warfarin dose prediction model [18]. Indeed, most studies on warfarin maintenance dose prediction demonstrate a large gap between the sample number of the patients in the medium-dose group and that in the low- and high-dose groups. It is worth noting that patients receiving low or high warfarin doses are more vulnerable to cardiovascular adverse events. Therefore, to improve the model prediction effect in low- and high-dose patient groups, we attempted to correct the CIL problem by adjusting the distribution structure of the training dataset before modeling.
Methods
Study Population
This was a retrospective multicenter study based on the Chinese Low-Intensity Anticoagulant Therapy after Heart Valve Replacement (CLIATHVR) database. The CLIATHVR database prospectively included all patients who underwent HVR and received warfarin treatment from June 1, 2012, to June 1, 2016, in 35 centers (15 provinces) in China. The study included adult patients who met the following criteria: (1) underwent HVR; (2) received warfarin to prevent valve-associated thrombosis after surgery; (3) conducted regular international normalized ratio (INR) monitoring; and (4) achieved a stable dose of warfarin (the INR value fluctuated by < 0.2 for three consecutive times under the fixed dose). The exclusion criteria were as follows: (1) patients in whom severe liver or kidney dysfunction occurred before or after surgery; (2) those who received a combination of other antiplatelet or anticoagulant agents or non-steroidal anti-inflammatory drugs; and (3) patients with embolism, bleeding events, or death during warfarin therapy (the absence of complications indicated the ideal statement under warfarin maintenance dose). All included patients provided informed consent for the procedure and data collection. The study protocol was approved by the Ethics Committee of West China Hospital of Sichuan University (ChiECRCT-201792).
Input and Output Variables
The input variables were selected in two ways: (1) clinical characteristics associated with the warfarin dose were chosen according to expert advice and published literature; and (2) preliminarily comprehensive screening was conducted to select potential variables related to the warfarin maintenance dose. The correlation coefficient matrix of input variables is developed, and the variables with strong collinearity are screened according to the variance inflation factor (VIF) of each variable (VIF > 10 was considered strong collinearity). The general linear model (GLM)-univariate method was further used to screen out variables based on both the level of statistical significance (P < 0.05) and η2 (> 0.002; where η2 is defined as the contribution of a specific input variable to the output variable). The output variable was the predicted warfarin maintenance dose, which was defined as the target value of the patients’ INR between 1.5 and 2.5 on at least three consecutive occasions.
Group Setting
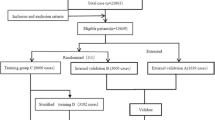
The entire dataset construction process is shown in Fig. 1. The eligible cases were divided into three groups as follows: the training set (80% of patients), the internal validation set (10% of patients), and the external validation set (the final 10% of patients by the enrollment time). We applied two training datasets with different structures to train the ANFIS model, in order to compare the prediction accuracy in the same model before and after preprocessing. The ANFIS model trained by the above training group (imbalanced training set) was called the imbalanced model, and the new dataset constructed by the method of equal random-stratified sampling from the imbalanced training set was called the balanced training set. Finally, the ANFIS model trained by this training set was called the balanced model. The purpose of the equal random-stratified sampling was to randomly sample the same number of cases in other groups according to the group containing the minimum number of cases; thus, the proportion of patients receiving high, medium, and low doses of warfarin in the balanced training set was 1:1:1.
Adaptive Neural-Fuzzy Inference System (ANFIS) Model
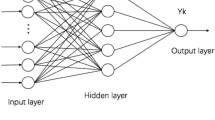
The ANFIS, as a classic machine learning algorithm, is a neuro-fuzzy system (NFS) that was proposed by Jang et al. in 1993 [19] and combines the advantages of a fuzzy inference system (FIS) and ANN. The ANFIS is driven by data and can automatically construct a set of if-then fuzzy rules, create an appropriate membership function, determine its parameters, and quickly form the mapping relationship between the input and output. Since the ANFIS has only limited parameter settings, it dramatically simplifies the problems of unclear and complicated characteristics in the process of system modeling. The models used in this study are based on the Takagi-Sugeno type of ANFIS. Figure 2 presents the overall structure of the ANFIS model. It includes a multi-layer feed-forward network with a total of five layers.
-
Layer 1 is the fuzzification layer in which each node represents a membership value to a linguistic term as a Gaussian function with the mean.
-
Layer 2 provides the strength of the rule using multiplication operator.
-
Layer 3 is the normalization layer which normalizes the strength of all rules according to the equation.
-
Layer 4 is a layer of adaptive nodes. Every node in this layer computes a linear function where the function coefficients are adapted by using the error function of the multi-layer feed-forward neural network.
-
Layer 5 is the output layer whose function is the summation of the net outputs of the nodes in layer 4.
This system is characterized by the back-propagation (BP) algorithm and least square algorithm, which determine the fuzzy rules and relevant parameters and automatically establish the mapping relationship between the input and output. The input of the model is the patient data, and the output is the predicted value of warfarin dosing.
The Prediction Ability of the Model
The primary outcomes were the mean difference between the predicted warfarin maintenance dose by the models and the actual dose in clinical practice and the predictive accuracy of the models that were evaluated by the internal and external validation sets with three indexes: the ideal predicted percentage, the mean absolute error (MAE), and the mean squared error (MSE). The ideal predicted percentage was defined as the percentage of patients whose predicted warfarin dose was within 20% of the actual dose. The MAE was the mean absolute difference between the predicted dosage and the actual dosage of warfarin. The MSE was the square of the difference between the two dosages. The warfarin dose was classed as low (< 1.875 mg/d), medium (1.875–3.125 mg/d), or high (> 3.125 mg/d).
Statistical Analyses
Microsoft Excel 2019 was used for data inclusion and preliminary screening of eligible cases. Categorical variables are expressed as number and percentage and were compared using the chi-square test. Continuous variables are expressed as mean with standard deviation (SD) and were compared using the paired Student’s t-test or Mann–Whitney U tests. MATLAB R2009a (MathWorks Inc, USA; code is presented in supplemental file) was used to establish the ANFIS models and predict the individual warfarin dose. The differences between the predicted dose and the actual dose of each ANFIS model were analyzed using the mean difference (< 0.3 mg/d considered to be an acceptable difference). The models’ predictive accuracy (MAE, MSE, ideal predicted percentage) was calculated using the ANFIS model results. Statistical analyses were performed using SPSS software, version 22.0 (SPSS Inc., Chicago, Illinois, U.S.A), with a P value < 0.05 indicating a significant difference.
Results
Data Acquisition and Variable Inclusion
As shown in Fig. 3, 19,595 patients who were enrolled in the CLIATHVR database between June 1, 2012, and June 1, 2016, were selected in this study. After eliminating 3424 unfinished cases and 726 cases that did not meet the criteria, 15,445 cases were eligible for further analysis. A further 337 cases containing missing data or abnormal values were excluded, and an original dataset, including 15,108 patients and consisting of 52 potential independent variables (patient characteristics, medical history, echocardiography indexes, preoperative laboratory results, surgical information, and postoperative warfarin medication information), was finally constructed. Of which, 8512 patients who were involved in our previous study [10] were also enrolled in the present study. Variables were selected based on the GLM univariate method (Table S1), and eight variables (including age, disease, weight, tricuspid valve disease, albumin level, creatinine level, usage of the first dose, and dosage of the first dose) were included for model construction (Table S2).
Datasets and Population Characteristics
Of 15,108 patients, 1511 were selected as the external validation set according to the admission time. From the remaining 13,597 patients, 1511 were selected as the internal validation set by random sampling, and 12,086 patients were selected as the imbalanced training set. The number of patients in the low-, medium-, and high-dose groups in the imbalanced training set was 1259 (10.42%), 9887 (81.81%), and 940 (7.78%), respectively, indicating a significant imbalance (imbalanced ratio, 10.5). Therefore, the equal random-stratified sampling method was used to construct a balanced training set that included 2820 patients (imbalanced ratio, 1.0). The baseline characteristics of the included variables in the different datasets are shown in Table 1. The mean age of the overall population was 50.84 ± 11.09 years, and the mean weight was 60.16 ± 10.77 kg. Warfarin was primarily used for the treatment of rheumatic heart disease (83.95%), with a mean maintenance dose of 2.65 ± 0.66 mg/d. The baseline characteristics were similar between the patients in the imbalanced training set and those in the balanced training set (P > 0.05 for each variable). The characteristics of the patients in the external validation set were significantly different from those of the patients in the other datasets (internal validation set, imbalanced training set, and imbalanced training set; P < 0.05 for weight, albumin level, creatinine level, usage of the first dose, dosage of the first dose, and warfarin maintenance dose), which satisfied the requirement for the external validation set.
ANFIS Model Construction
On the basis of the eight variables as the input layer and the warfarin maintenance dose as the output layer, the imbalanced and the balanced training sets were used to train the original ANFIS model, respectively. After self-adjustment of the model with default settings, an imbalanced model with two fuzzy rules and a balanced model with four fuzzy rules were constructed (Fig. S1). Meanwhile, the individual variable’s membership functions and the warfarin maintenance dose are shown in Fig. S2.
The Overall Prediction Ability of Models
As shown in Table 2, the predictive warfarin doses were close to the actual doses, with a low mean difference (< 0.3 mg/d for each model). The overall prediction abilities of the imbalanced model and the balanced model are summarized in Table 3. For internal validation, the overall prediction accuracy of the balanced ANFIS model was 75.31%, and that of the imbalanced ANFIS model was 78.16%. The difference in the overall prediction accuracy between the two models was not statistically significant (P = 0.064). The MAE of the balanced model and the imbalanced model was 0.421 and 0.368, respectively. The MSE of the balanced model was 0.433, and that of the imbalanced model was 0.388. For external validation, the overall prediction accuracy of the balanced ANFIS model and the imbalanced model was 73.46% and 74.39%, respectively, with no significant difference (P = 0.562). The MAE of the balanced model was 0.422, and that of the imbalanced model was 0.370. The MSE of the balanced model was 0.413, and that of the imbalanced model was 0.386.
The Prediction Ability of the Two Models in Different Warfarin Dose Groups
The prediction difference of models across warfarin doses (low-dose, < 1.875 mg/d; medium-dose, 1.875–3.125 mg/d; high-dose, > 3.125 mg/d) is outlined in Fig. 4 and Table S3. As for internal validation, although the balanced model lowered the prediction accuracy by 6.21% compared with the imbalanced model in the medium-dose group (P < 0.001), it inversely significantly increased the prediction accuracy in the low- and high-dose groups (P < 0.001 and P = 0.047, respectively). Similar results were found in the external validation set, which strengthened the conclusion that the balanced ANFIS model could improve the prediction effect in the low- and high-dose groups.
Discussion
Major Findings
In this study, we simultaneously constructed two ANFIS models, namely, the imbalanced model and the balanced model, to predict the warfarin maintenance dose, based on a retrospective multicenter database involving 35 centers and 15,108 patients after HVR. The major findings were as follows: (I) the imbalanced ANFIS model, based on a training set of 12,086 cases, could accurately predict the warfarin maintenance dose for Chinese patients undergoing HVR, with an ideal prediction percentage of 74.39–78.16%, MAE of 0.37 mg/daily, and MSE of 0.39 mg/daily; (II) the balanced ANFIS model that used equal random-stratified sampling and was based on a training set of 2820 cases also achieved an accurate prediction property of warfarin maintenance dose (ideal prediction percentage, 73.46–75.31%; MAE, 0.42 mg/daily; MSE, 0.43 mg/daily); (III) compared to the imbalanced model, the balanced model had a significantly higher prediction accuracy in the low-dose warfarin group (internal validation, 14.46% vs. 3.01%; P < 0.001) and the high-dose warfarin group (34.71% vs. 23.14%; P = 0.047); (IV) the results of external validation were in line with the results of internal validation, thus strengthening the conclusion that the ANFIS model could improve the model prediction effect.
Comparison with Previous Models
Table S4 summarizes the current warfarin prediction models. In 2004, Gage et al. firstly created a warfarin dosage prediction model based on 369 patients [20]. This study explored eight variables (age, sex, body surface area [BSA], race, amiodarone, simvastatin use, INR, CYP2C19) using an MLR model and achieved a 39% predictive ability to explain the variance of the warfarin maintenance dose. Of note, the CYP2C9*2 and CYP2C9*3 alleles contributed to a dominating weight in the said model. Since then, six further studies have been conducted in order to gain a higher predictive accuracy of the model in a Caucasian population [2, 3, 21,22,23,24]. Although these studies achieved considerable predictive abilities (R2, 47–73%) through involving certain pharmacogenomic information (e.g., CYP2C19, VKORC1, GGCX), they had two main limitations: the small sample sizes (< 350 patients), which limited the representation of the population; and a lack of external validation, which limited the extrapolation of models to large patient populations in real-world practice. In 2008, Gage et al. developed another pharmacogenetic algorithm based on 1015 patients and nine predictors (age, BSA, smoking, race, amiodarone use, current thrombosis, CYP2C9, VKORC1, target INR) [7]; this model could explain 53–54% of the variability in the warfarin dose in the derivation and validation cohorts. Furthermore, a nonprofit website was developed to facilitate the use of this pharmacogenetic and clinical equation (www.WarfarinDosing.org). The following year, the IWPC created a novel pharmacogenetic algorithm that included 4043 patients from 21 various research groups in nine countries and eight factors (age, weight, height, race, amiodarone status, enzyme inducers, CYP2C9, and VKORC1) [6]. This model could explain 43–47% of the variability in the derivation and validation populations and provided accurate dose estimates, as evidenced by a low MAE (8.3 mg/week). In addition, the differences in the performance of the model in the low-dose (≤ 21 mg/week), medium-dose (21–49 mg/per week), and high-dose (≥ 49 mg/week) groups were evaluated. Although the Gage and IWPC models have addressed the above limitations, it may not be appropriate to directly extrapolate these results for a Chinese population due to the variation in warfarin sensitivity across ethnic groups (weight, dietary habit, drug interaction, genotype, adherence, etc.). All of these inherent issues have fueled the development of warfarin prediction models for the Chinese population. However, the current Chinese medical insurance coverage only covers genetic testing for warfarin dosage prediction for patients with a high risk of bleeding or labile INR values, which is a barrier to its utilization. Considering the latter limitation, the current models conducted for a Chinese population have included small sample sizes combining both clinical and pharmacogenomic variables, which limited the generalizability of models [25,26,27,28,29,30,31]. Therefore, developing the optimal prediction model for warfarin dose based on explicable clinical variables is challenging.
The MLR method presents certain irreconcilable issues such as poor behavior of the non-linear relationship between variables; thus, the MLR is unlikely to be an optimal method for predicting the warfarin dose [32]. Recently, several artificial intelligence modeling technologies, including support vector machines and a general regression neural network, have been used for warfarin dosage predication [33, 34]; however, these models showed a relatively low predictive ability (< 50% of the ideal predicted percentage). Our study team has made numerous attempts in warfarin model development and achieved a 63% predictive accuracy based on BPGA and ANFIS models [10, 12, 35]. This study further included 15,108 patients who underwent HVR from 35 centers and used balanced training set preprocessing with the equal random-stratified sampling method. Compared with the results of the IWPC model, both the imbalanced and the balanced ANFIS models had better performance in terms of ideal prediction percentage (73.46–74.39% for ANFIS vs. 45.5% for IWPC) and MAE (2.59–2.95 mg/week for ANFIS vs. 8.5 mg/week for IWPC) in external validation cohorts. Hence, the ANFIS method based on big data is a feasible and optimal modeling technology to improve the prediction ability for estimating the warfarin maintenance dose.
Reasons for Improved Prediction Property in Low- and High-Dose Groups
Patients receiving low or high warfarin doses are more vulnerable to thromboembolic and bleeding events due to difficulty with INR control. To date, no study has been specifically designed to address this concern. Our previous studies found an extremely low prediction accuracy in the low-dose group (0.0% by BPNN [13] and 9.1% by ANFIS [10]) and high-dose group (0.0% by BPGA [12]). Considering the distribution of patients across different doses in the training set, the proportion in the medium-dose group was higher than that in the low- and high-dose groups (low-dose, 10.41%; medium-dose, 81.81%; high-dose, 7.78%). This explains why our previous models showed better performance in the medium-dose group but poor performance in the low-dose group. It is known as the CIL problem. In brief, because the sample size of one class is considerably larger than that of the other classes, the machine learning models detect more characteristics of the people in the bigger class than those of the people in the smaller class; this leads to insufficient data learning of smaller-scale categories, resulting in an unsatisfactory prediction effect of the model in a smaller class [36]. We used the equal random-stratified sampling method to address this problem, which can balance the number of patients in each group through random sampling [37]. Using the balanced training set, the model results indicated an increased prediction accuracy compared to the imbalanced model (low-dose, 14.46–24.34% vs. 3.01–3.62%; high-dose, 29.58–34.71% vs. 21.12–23.14%).
Clinical Relevance
When lacking genetic information in a clinical setting, this ANFIS method could provide high accurate warfarin dose estimates on the basis of clinical variables (including age, disease, weight, tricuspid valve disease, albumin level, creatinine level, usage of the first dose, and dosage of the first dose). This could aid physicians and pharmacists in the selection of patients who will likely be suited to low or high doses of warfarin, thus allowing earlier and more aggressive intervention to control INR.
Strengths and Limitations
The main strengths of this study were as follows: first, this study used a large sample of 15,108 Chinese patients from 35 centers who received warfarin after HVR to develop and validate the models; second, we applied the equal random-stratified sampling method to address the CIL problem that resulted in the low predicted ability in the low- and high-dose groups; and third, we validated the models using both internal and external validation cohorts. However, this study also had some limitations. First, this was a retrospective study that may have a certain selection bias. Second, some of the possible determinants of warfarin dose, such as diet information and patient genotypes (CYP2C9 and VKORC1), are not available in our study, which may limit the performance of the models. Third, clinical adverse events related to warfarin use were not examined in this study. Given the above limitations, further prospective studies with more potential predictors need to be carried out to further improve the model performance using machine learning techniques.
Conclusions
This study constructed two ANFIS models to predict the warfarin maintenance dose based on 15,108 patients who underwent HVR from 35 centers. The results showed that both imbalanced and balanced models could accurately predict warfarin maintenance dose (ideal prediction percentage > 70%). Besides, the balanced model contributed to improved prediction ability in the low- and high-dose warfarin groups.
Data Availability
All data can be obtained by contacting the corresponding author.
References
Kirley K, Qato DM, Kornfield R, Stafford RS, Alexander GC. National trends in oral anticoagulant use in the United States, 2007 to 2011. Circulation-Cardiov Qual Outcomes. 2012;5(5):615–21.
Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116(22):2563–70.
Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106(7):2329–33.
Jorgensen AL, Pirmohamed M. Risk modeling strategies for pharmacogenetic studies. Pharmacogenomics. 2011;12(3):397–410.
Yan H, Yin J-Y, Zhang W, Li X. Possible strategies to make warfarin dosing algorithm prediction more accurately in patients with extreme dose. Clin Pharmacol Therap. 2018;103(2):184.
Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MTM, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data (vol 360, pg 753, 2009). N Engl J Med. 2009;361(16):1613.
Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, Ridker PM, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clinical Pharmacol Therap. 2008;84(3):326–31.
Anderson JL, Horne BD, Stevens SM, Woller SC, Samuelson KM, Mansfield JW, et al. A randomized and clinical effectiveness trial comparing two pharmacogenetic algorithms and standard care for individualizing warfarin dosing (CoumaGen-II). Circulation. 2012;125(16):1997.
Li X, Liu R, Luo Z-Y, Yan H, Huang W-H, Yin J-Y, et al. Comparison of the predictive abilities of pharmacogenetics-based warfarin dosing algorithms using seven mathematical models in Chinese patients. Pharmacogenomics. 2015;16(6):583–90.
Tao H, Li Q, Zhou Q, Chen J, Fu B, Wang J, et al. A prediction study of warfarin individual stable dose after mechanical heart valve replacement: adaptive neural-fuzzy inference system prediction. BMC Surg. 2018;18:10.
Zhou Q, Kwong J, Chen J, Qin W, Chen J, Dong L, et al. Use of artificial neural network to predict warfarin individualized dosage regime in Chinese patients receiving low-intensity anticoagulation after heart valve replacement. Int J Cardiol. 2014;176(3):1462–4.
Li Q, Tao H, Wang J, Zhou Q, Chen J, Qin WZ, et al. Warfarin maintenance dose prediction for patients undergoing heart valve replacement-a hybrid model with genetic algorithm and back-propagation neural network. Sci Rep. 2018;8:9712.
Li Q, Wang J, Tao H, Zhou Q, Chen J, Fu B, et al. The prediction model of warfarin individual maintenance dose for patients undergoing heart valve replacement, based on the back propagation neural network. Clin Drug Investig. 2020;40(1):41–53.
Groszek B, Piszczek P. Vitamin K antagonists overdose. Przegl Lek. 2015;72(9):468–71.
Heffler E, Campisi R, Ferri S, Crimi N. A bloody mess: an unusual case of diffuse alveolar hemorrhage because of warfarin overdose. Am J Ther. 2016;23(5):E1280–E3.
Levine M, Pizon AF, Padilla-Jones A, Ruha A-M. Warfarin overdose: a 25-year experience. J Med Toxicol. 2014;10(2):156–64.
Wang SV, Franklin JM, Glynn RJ, Schneeweiss S, Eddings W, Gagne JJ. Prediction of rates of thromboembolic and major bleeding outcomes with dabigatran or warfarin among patients with atrial fibrillation: new initiator cohort study. Bmj-British Med J. 2016;353. https://doi.org/10.1136/bmj.i2607.
Zhang Y, Liu B, Cai J, Zhang S. Ensemble weighted extreme learning machine for imbalanced data classification based on differential evolution. Neural Comput Applic. 2017;28:S259–67.
Jang JR, Sun CT. Functional equivalence between radial basis function networks and fuzzy inference systems. IEEE Trans Neural Netw. 1993;4(1):156–9.
Gage BF, Eby C, Milligan PE, Banet GA, Duncan JR, McLeod HL. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb Haemost. 2004;91(1):87–94.
Aquilante CL, Langaee TY, Lopez LM, Yarandi HN, Tromberg JS, Mohuczy D, et al. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P4502C9 gene polymorphisms on warfarin dose requirements. Clinical Pharmacol Therap. 2006;79(4):291–302.
Herman D, Peternel P, Stegnar M, Breskvar K, Dolzan V. The influence of sequence variations in factor VII, gamma-glutamyl carboxylase and vitamin K epoxide reductase complex genes on warfarin dose requirement. Thromb Haemost. 2006;95(5):782–7.
Wadelius M, Chen LY, Eriksson N, Bumpstead S, Ghori J, Wadelius C, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007;121(1):23–34.
Zhu Y, Shennan M, Reynolds KK, Johnson NA, Herrnberger MR, Valdes R Jr, et al. Estimation of warfarin maintenance dose based on VKORCI (-1639 G > A) and CYP2C9 genotypes. Clin Chem. 2007;53(7):1199–205.
Miao L, Yang J, Huang C, Shen Z. Contribution of age, body weight, and CYP2C9 and VKORC1 genotype to the anticoagulant response to warfarin: proposal for a new dosing regimen in Chinese patients. Eur J Clin Pharmacol. 2007;63(12):1135–41.
Wen MS, Lee MTM, Chen JJ, Chuang HP, Lu LS, Chen CH, et al. Prospective study of warfarin dosage requirements based on CYP2C9 and VKORC1 genotypes. Clinical Pharmacol Therap. 2008;84(1):83–9.
Huang S-W, Chen H-S, Wang X-Q, Huang L, Xu D-L, Hu X-J, et al. Validation of VKORC1 and CYP2C9 genotypes on interindividual warfarin maintenance dose: a prospective study in Chinese patients. Pharmacogenet Genomics. 2009;19(3):226–34.
Cen H-J, Zeng W-T, Leng X-Y, Huang M, Chen X, Li J-L, et al. CYP4F2 rs2108622: a minor significant genetic factor of warfarin dose in Han Chinese patients with mechanical heart valve replacement. Br J Clin Pharmacol. 2010;70(2):234–40.
You JHS, Wong RSM, Waye MMY, Mu Y, Lim CK. Choi K-c et al. Warfarin dosing algorithm using clinical, demographic and pharmacogenetic data from Chinese patients. J Thromb Thrombolysis. 2011;31(1):113–8.
Wei M, Ye F, Xie D, Zhu Y, Zhu J, Tao Y, et al. A new algorithm to predict warfarin dose from polymorphisms of CYP4F2, CYP2C9 and VKORC1 and clinical variables: derivation in Han Chinese patients with non valvular atrial fibrillation. Thromb Haemost. 2012;107(6):1083–91.
Lou Y, Liu H, Han L, Xie S, Huang YL, Li YS. The study of warfarin maintenance dose algorithm in Chinese Han population. Int J Cardiol. 2013;163:S9.
Liu R, Li X, Zhang W, Zhou HH. Comparison of nine statistical model based warfarin pharmacogenetic dosing algorithms using the racially diverse international warfarin pharmacogenetic consortium cohort database. PLoS One. 2015;10(8):e0135784.
Tao Y, Chen YJ, Fu X, Jiang B, Zhang Y. Evolutionary ensemble learning algorithm to modeling of warfarin dose prediction for Chinese. IEEE J Biomed Health Inform. 2019;23(1):395–406.
Tao Y, Chen YJ, Xue L, Xie C, Jiang B, Zhang Y. An ensemble model with clustering assumption for warfarin dose prediction in Chinese patients. IEEE J Biomed Health Inform. 2019;23(6):2642–54.
Li S, Garcia DA, Khorana AA, Carrier M, Lyman GH, Kalmanti L, et al. Treatment of vascular thrombosis in antiphospholipid syndrome: an update. Cancer. 2020;40(1):31–7.
He H, Garcia EA. Learning from imbalanced data. IEEE Trans Knowl Data Eng. 2009;21(9):1263–84.
Hirzel A, Guisan A. Which is the optimal sampling strategy for habitat suitability modelling. Ecol Model. 2002;157(2-3):331–41.
Funding
This study was supported by the National Natural Science Foundation of China (71974137 and 81641021), research funds of Shanghai Health and Family Planning commission (20184Y0022), cultivation fund of clinical research of Renji Hospital (PY2018-III-06), Clinical Pharmacy Innovation Research Institute of Shanghai Jiao Tong University School of Medicine (CXYJY2019ZD001), and Shanghai “Rising Stars of Medical Talent” Youth Development Program — Youth Medical Talents — Clinical Pharmacist Program (SHWJRS (2019)_072).
Author information
Authors and Affiliations
Contributions
Chen is the guarantor of the entire manuscript. Gu and Huang contributed to the study conception and design, critical revision of the manuscript for important intellectual content, and final approval of the published version. Li, Zhou, Wang, and Fu contributed to data acquisition, analysis, and interpretation.
Corresponding author
Ethics declarations
Ethical Statement
This study was registered in the Chinese Clinical Trial Register platform (trial number, ChiCTR-OCH-10001185). The study protocol was approved by the Ethics Committee of West China Hospital of Sichuan University (ChiECRCT-201792). All participants signed written informed consent.
Conflicts of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOC 718 kb)
Rights and permissions
About this article
Cite this article
Gu, ZC., Huang, SR., Dong, L. et al. An Adapted Neural-Fuzzy Inference System Model Using Preprocessed Balance Data to Improve the Predictive Accuracy of Warfarin Maintenance Dosing in Patients After Heart Valve Replacement. Cardiovasc Drugs Ther 36, 879–889 (2022). https://doi.org/10.1007/s10557-021-07191-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10557-021-07191-1