Abstract
Polymerase chain reaction (PCR) has become a powerful tool for detecting various diseases due to its high sensitivity and specificity. However, the long thermocycling time and the bulky system have limited the application of PCR devices in Point-of-care testing. Herein, we have proposed an efficient, low-cost, and hand-hold PCR microdevice, mainly including a control module based on water-cooling technology and an amplification module fabricated by 3D printing. The whole device is tiny and can be easily hand-held with a size of about 110 mm × 100 mm × 40 mm and a weight of about 300 g at a low cost of about $170.83. Based on the water-cooling technology, the device can efficiently perform 30 thermal cycles within 46 min at a heating/cooling rate of 4.0/8.1 ℃/s. To test our instrument, plasmid DNA dilutions were amplified with this device; the results demonstrate successful nucleic acid amplification of the plasmid DNA and exhibit the promise of this device for Point-of-care testing.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The analysis of nucleic acids plays an irreplaceable role in detecting various viruses and bacteria with sensitivity and specificity advantages (Chan et al. 2016). To detect nucleic acids, amplification is always needed to improve the concentration of nucleic acids extracted from samples. The polymerase chain reaction (PCR) has become a sought-after way to achieve nucleic acid amplification for detection (Zhu et al. 2020a, b), which can exponentially amplify nucleic acids and be applied in various detection scenes, including food safety supervision (Zhang et al. 2010), environmental conservation (Hutchins et al. 2018), genetic disorders diagnosis (Camunas-Soler et al. 2018), cancer detection (Li et al. 2018).
PCR technology has been quickly developed in recent years due to its excellent prospects in the biological and biomedical fields. Nevertheless, there are several shortcomings of the PCR technology in conventional laboratories (Liao et al. 2016), such as using sophisticated instruments, cumbersome and time-consuming procedures, and requiring professional operators. For the convenient use of PCR technology, some commercial PCR devices have also been proposed and applied to detect diseases. But the drawbacks of bulk and high cost are not overcome completely (Pan et al. 2020), which hinders the extended use of PCR technology, especially for Point-of-care testing. Thus, developing small, low-cost, and portable PCR devices is critical to perform more practical and convenient analyses of nucleic acids (Ahrberg et al. 2016).
To address this issue, many researchers have focused on enhancing PCR devices’ integration level, portability, and efficiency. For example, Jyh et al. (Chen et al. 2018) proposed a nucleic acids amplification device using a microfluidic chip; it can amplify a 190-bp segment of Bartonella DNA. A more integrated system (Kopparthy et al. 2020) has also been developed, enabling simultaneous amplification and analysis of samples, but the syringe pump used for driving PCR reagents is not practical. Roberto et al. (Mendoza-Gallegos et al. 2018) proposed a more portable and functional real-time PCR device made of 3D-printed parts and ready-made electronics at $193.2; however, the total time required for a 40-cycle was ~ 120 min, which is not conducive for rapid detections. To improve the efficiency of PCR reactions, efficient heating/cooling methods are necessary for the design of PCR devices. Notwithstanding, researchers have applied various heating technologies to PCR devices and obtained remarkable results (Li et al. 2019; Shan et al. 2020; Pak et al. 2012; Yang et al. 2022; Tzivelekis et al. 2021; Cho et al. 2013), the cooling process during PCR reactions was frequently achieved with a cooling fan (Zhu et al. 2020a, b; Xu et al. 2021; Gorgannezhad et al. 2019; Zhou et al. 2019) which is bulky and mediocre in cooling rate.
Compared to cooling fans, liquid cooling is a more efficient way for heat dissipation, which has also been applied to PCR systems (Chen et al. 2013a, b). For example, Chen et al. proposed a continuous-flow PCR chip by water cooling, in which the temperature zones were controlled by two thermal controllers and a cooling channel (Chen et al. 2013a, b). However, the water-cooling technology was used to form the temperature zone for the annealing of DNA but not to improve the efficiency of the PCR reaction. Recently, Alihosseini et al. reported the effect of liquid cooling on PCR performance with different cross-section shapes of microchannels, but this is limited to numerical simulations (Alihosseini et al. 2021).
In such a context, we proposed a PCR microdevice that innovatively uses water as the temperature transfer medium to speed up PCR reactions. The device consists of two main parts: a temperature control module and a nucleic acids amplification module. It can enable a rapid cooling rate (8.0 ℃/s) during the PCR reaction and shorten the duration of the cooling process in each thermal cycle. To simplify the fabrication of the PCR microdevice, we use a 3D printed frame, a plastic pedestal, and off-the-peg components to assemble the microdevice at a low cost of $170.83, and it can be easily carried with a total size of 110 mm × 100 mm × 40 mm and the weight of 300 g. Finally, we accomplished the detection of plasmid DNA with a total time of 46 min for 30 cycles. Therefore, given the advantages of low cost, high portability, and efficient amplification, the proposed PCR microdevice based on water-cooling technology is beneficial for analyzing nucleic acids in households or remote regions.
2 Experimental
2.1 Materials and instruments
The Peltier heater (CP1140203) used in this study was sourced from DigiKey Electronics, USA, with a size of 40 mm × 20 mm × 3 mm and a maximum working voltage of 7.6 V. The micro water pump (D200) used for driving water was sourced from RS PRO, UK, with a size of 42 mm × 27 mm × 17 mm and a maximum working voltage of 4.5 V. The amplification module used for the PCR reaction was fabricated by the 3D printer (Form 3+, Formlabs, USA) with the matched clear resin (Formlabs, USA). The aluminum heat sink (a17041100ux1232) used to cool the heater was sourced from Sourcingmap, China, with a size of 40 mm × 40 mm × 11 mm. To assemble the whole PCR microdevice, the 3D printer (Ultimaker S5, Ultimaker, Netherlands) was used to print the frame of the entire device and the pedestal of the amplification module (designed by SOLIDWORKS 2019 software) with the matched white PLA material (Ultimaker, Netherlands). The thermal imaging camera (optris PI450, with a resolution of 382 × 288) used to measure the surface temperature of the amplification module was sourced from optris, Germany. The soft silicon tube (2 mm × 3 mm of inner and outer diameter) used to connect the pump with the amplification module was sourced from Sourcingmap, China.
To control the temperature precisely, we made a control module based on a PCB board (9 mm × 4 mm) and prepared the control program of the module with the open-source software platform (Arduino, Italy). All the components of the control module and their details are as follows: microcontroller (Adafruit Feather HUZZAH ESP8266, Adafruit, USA), type-K thermocouple (110–4482, RS PRO, UK), thermocouple amplifier (MAX31855, Adafruit, USA), MOSFET (IRL540N, International Rectifier, USA), regulator (AMS1117, AMS, Austria, 5 V to 1.5 V), 0.96-inch OLED display (Adafruit, USA), PCB board (Adafruit, USA). The power supply used to power the whole device was sourced from RS PRO, UK, with an output of 5 V.
To test the proposed PCR microdevice, an amplification of plasmid DNA from Escherichia coli was performed. The template DNA (50 µg/mL), forward primer (10 µM), and reverse primer (10 µM) used in the amplification were sourced from GENEWIZ, USA. The 5X Q5 reaction buffer, 5X Q5 high GC enhancer, and Q5 high-Fidelity DNA polymerase were sourced from New England Biolabs (Beijing) LTD, China. The dNTP (2 mM) was sourced from ThermoFisher Scientific, USA. The mineral oil used in the PCR reaction was sourced from Beyotime Biotechnology Co., Ltd, China. The agarose used in this study was sourced from Aladdin Biochemical Technology Co., Ltd, China. The gel imaging system (Tanon 1600) was sourced from Shanghai Tianneng Life Science Co., LTD, China.
2.2 System design and instrumentation
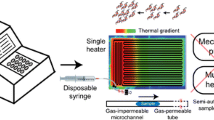
A schematic drawing of the hand-hold PCR microdevice based on water-cooling technology is shown in Fig. 1a, including an amplification module based on water-cooling and a control module. The plastic pedestal for the amplification module and the frame for the whole device was fabricated by 3D printing. All components were tightly assembled to form a hand-hold microdevice of approximately 110 mm × 100 mm × 40 mm in size and 300 g in weight, as shown in Fig. 1b, which can be easy to carry and applied to in-situ detection. The interior photograph of the PCR microdevice is performed in Fig. 1c to show more details of the device. Furthermore, the temperature of the PCR reaction reagent and the cycle number of the PCR reaction can be real-timely displayed in an OLED display fixed on the top side of the microdevice.
Due to the cost-effective off-the-peg components and 3D printing method, the entire device only costs about $170.83, which mainly includes the control module ($72.99), water pump ($45.23), Peltier heater ($29.10), and heat sink ($1.37). Without including the cost of 3D printing material, the whole cost of our device is highly affordable compared to that of commercial PCR instruments (e.g., THG-96, about $1658).
2.3 Control module
The schematic diagram of the control module used to control the temperature automatically is shown in Fig. 2. The heater and the micro water pump were used for heating and cooling PCR reagents, respectively, which are cooperatively controlled by the microcontroller and MOSFETs. To reduce the temperature deviation between the actual value and the preset value, the proportional integration derivative (PID) algorithm was used to control the heater. A type-K thermocouple connected to a thermocouple amplifier was used to monitor the temperature of the PCR reagent in real-time. Then the temperature and the cycle number can be real-timely displayed in an OLED display connected to the microcontroller. Finally, the whole device was powered by a portable switching power supply of 5 V that can easily plug into the power socket in the home, and it can also be replaced with a portable battery with similar power capacity.
2.4 Amplification module
For the amplification of nucleic acids, an amplification module was fabricated by a 3D printer; the schematic diagram of the module is shown in Fig. 3a. In this module, there is a microchannel (about 12 mm × 15 mm in size) for water cooling, an inlet, and an outlet (1 mm × 3 mm of inner and outer diameter) for driving water in the microchannel; there are also four chambers with the diameter of 2 mm and per chamber can hold about 3.14 µl of PCR reagent; thus, this PCR microdevice can perform the analyses in parallel using multiple reagents or different genes. Figure 3b shows the photo of the amplification module fabricated with 3D printing. Figure 3c and d are the schematic diagrams of the heating/cooling process during the PCR reaction, the oil layer above the chambers was used to avoid the evaporation of the PCR reagents. To achieve temperature control, the bottom surface of the amplification module was aligned and directly touched with the heater to accomplish thermal conduction; the microchannel was connected to the micro-pump to fulfill thermal convection. Therefore, the PCR reagent will be heated while the working states of the pump and the heater are off and on, and will be cool while the working states are reversed.
3 Results and discussion
To ensure successful nucleic acid amplification, the temperature of the reaction chambers should be consistent with the set value, and the heat distribution in these chambers should be as homogeneous as possible to make them all work. Thus, an infrared thermal imaging camera (optris PI 450, optris, with a resolution of 382 × 288) was used to monitor and evaluate the temperature of these chambers. Figure 4a is a thermograph of the amplification module. We also counted the frequency distribution of the temperature in the reaction chambers at working temperatures of the PCR reaction (58 ℃, 72 ℃, and 98 ℃), as shown in Fig. 4b. The averages of temperature at 58 ℃, 72 ℃, and 98 ℃ are approximately 57.89 ℃, 72.95 ℃, and 97.56 ℃, respectively; the standard deviations of these three temperatures are approximately 1.07%, 1.12%, and 0.94%, respectively, showing good heat distribution.
Since the rates of heating and cooling have a significant influence on the whole period, we assessed the performance of our device from the flow rate of water, the thickness of water layer, and the thickness of polymer wall; the meaning of each parameter is illustrated in Fig. 5a. All the heating (58℃ to 98 ℃) and cooling (98℃ to 58 ℃) processes during the test are similar to the real temperature change during PCR reaction, the type-K thermocouple (with a probe diameter of 1 mm) was put into a reaction chamber to measure temperature and then calculating the heating/cooling rates. First, we evaluate the cooling rate under different flow rates of water (Fig. 5b); it shows that the flow rate of water has a minor influence on the cooling rate, which means the increased flow rate has little influence on the thermal convection in the amplification module due to the poor coefficient of thermal conductivity of polymer material. The heating/cooling rates of the reaction chamber with different thicknesses of polymer wall were tested and shown in Fig. 5c; it shows that the heating/cooling rates are enhanced markedly with thinner polymer wall, this is because the thinner polymer wall led to greater heat flux. Figure 5d shows the relationship between heating/cooling rates and the thickness of water layer, as the thickness of water layer increases, the heating rate decreases and the cooling rate increases; it can be attributed to the thicker water layer can absorb more heat from the heater and reaction chambers during the heating and cooling processes, respectively. These results indicate that the key to speeding up the PCR reaction in the proposed microdevice is to reduce the thickness of polymer wall as much as possible.
Heat/cooling performance of amplification module based on water cooling. (a): schematic diagram of different parameters in the amplification module. (b): comparison of cooling rates of a chamber with different flow rates of water. (c): comparison of heating and cooling rates of a chamber with different thicknesses of polymer wall. (d): comparison of heating and cooling rates of a chamber with different thicknesses of water layer
For polymer-based PCR devices, the poor coefficient of thermal conductivity limits the efficiency of PCR reactions. The performance of some typical polymer PCR devices is listed in Table 1 for comparison with the microdevice in this research. It can be found that the device in this study can achieve a more rapid cooling process than the method of cooling fan or natural cooling with the advantages of low cost and miniaturization. Even though the heating rate of this study is not prominent, it can be further promoted by using a more powerful heater.
To test the proposed PCR microdevice, a real PCR protocol was implemented, which include 30 s at 98 ℃ for pre-degeneration, followed by 30 cycles of 98 ℃ 10 s for denaturing 58 ℃ 30 s for annealing, and 72 ℃ 30 s for extending. For fully extending, we adjusted the extending time to 2 min in the last cycle. The parameters of the amplification module used in the PCR protocol are 200 μm (the thickness of the polymer wall), 1 mm (the thickness of the water layer), and 1.5 ml/s (the flow rate of water). During the PCR protocol, the temperature trace was monitored in one of the chambers of the amplification module with the thermocouple, as shown in Fig. 6. The total time required for a completed PCR reaction is about 46 min (Fig. 6a), and about 85 s for a single cycle (Fig. 6b). The average heating/cooling rates are about 4.0/8.1 ℃/s with a PID algorithm to control the temperature automatically. It indicates that a more efficient PCR reaction can be fulfilled with our device than that of commercial PCR devices (e.g., T-100 Thermal Cycler, BIO-RAD, USA) which need 80 min for the same PCR reaction.
An amplification of plasmid DNA of 369 base pairs was performed on both the proposed microdevice and a commercial thermocycler (T-100 Thermal Cycler, BIO-RAD, USA) to verify our device. The reaction solution was prepared in quantities of 25 µL with 5 µL 5X Q5 reaction buffer, 2.5 µL dNTP (2 mM), 1.25 µL forward primer (10 µM), 1.25 µL reverse primer (10 µM), 0.5 µL Template DNA (50 µg/mL), 0.25 µL Q5 high-Fidelity DNA polymerase, 5 µL 5X Q5 high GC enhancer, and 9.25 µL ddH2O. The nucleotide sequences of the DNA template and primers used in this study are performed in Table 2. Each chamber of the proposed device was loaded with 2.5 µL PCR reaction solution, and the commercial thermocycler was loaded with 10 µL PCR reaction solution for comparison. After running 30 thermal cycles, the products of both devices were collected and verified by the electrophoresis method simultaneously.
Figure 7 shows the gel-electrophoresis-based amplification results of the plasmid DNA with both the proposed microdevice and the commercial thermocycler. In the negative control, DI water with the same volume was used instead of the template DNA. Visible bands appeared in lane 2 (amplifying with the commercial thermocycler) and lanes 3–5 (amplifying three times with the proposed microdevice), demonstrating the successful DNA amplification achievable with the proposed PCR microdevice. In addition, the similar results of lanes 3–5 show the stable performance of our device.
4 Conclusion
This study proposed an efficient, low-cost, and hand-hold PCR microdevice based on water-cooling technology for nucleic acid amplification. The whole device is approximately 110 mm × 100 mm × 40 mm in size and 300 g in weight, which is about 63 times smaller than a commercial PCR device (e.g., T-100 Thermal Cycler, BIO-RAD, USA). In addition, the PCR microdevice has a low cost of about $170.83 due to it was made of off-the-peg components, a 3D printed frame, and a plastic pedestal. With this PCR microdevice, a rapid thermal cycle of 4.0/8.1 ℃/s of heating/cooling rates can be fulfilled, which is faster than most research, especially in cooling rate. For the demonstration of the proposed microdevice, a nucleic acid amplification of plasmid DNA of 369 base pairs was successfully performed with this microdevice.
Compared with conventional PCR devices based on cooling fans or passive cooling, this device can achieve efficient PCR reactions with high portability and low cost. Furthermore, the fabrication of this device is not complicated and there is no need to use sophisticated equipment. It is a promising tool for Point-of-care testing, such as infectious disease detection.
Data Availability
The date and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
K. Chan, S.C. Weaver, P.-Y. Wong, S. Lie, E. Wang, M. Guerbois, S.P. Vayugundla, S. Wong, Rapid, Affordable and Portable Medium-Throughput Molecular device for Zika Virus. Sci. Rep 6(1), 38223 (2016)
H. Zhu, H. Zhang, Y. Xu, S. Laššáková, M. Korabečná, P. Neuzil, PCR past, present and future. BioTechniques 69 (2020a)
W. Zhang, Z. Xie, H. Zuo, X. Ding, X. Pei, Detection of food-borne pathogens with polymerase chain reaction and introduction of food safety supervision system in China. Qual. Assur. Saf. Crops Foods 2(1), 13–21 (2010)
P.R. Hutchins, A.J. Sepulveda, R.M. Martin, L.R. Hopper, A probe-based quantitative PCR assay for detecting Tetracapsuloides bryosalmonae in fish tissue and environmental DNA water samples. Conserv. Genet. Resour. 10(3), 317–319 (2018)
J. Camunas-Soler, H. Lee, L. Hudgins, S.R. Hintz, Y.J. Blumenfeld, Y.Y. El-Sayed, S.R. Quake, Noninvasive prenatal diagnosis of single-gene Disorders by Use of Droplet Digital PCR. Clin. Chem 64(2), 336–345 (2018)
T. Li, Y. Shao, L. Fu, Y. Xie, L. Zhu, W. Sun, R. Yu, B. Xiao, J. Guo, Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J. Mol. Med. (Berl) 96(1), 85–96 (2018)
S.C. Liao, J. Peng, M.G. Mauk, S. Awasthi, J. Song, H. Friedman, H.H. Bau, C. Liu, Smart Cup: A Minimally-Instrumented, Smartphone-Based Point-of-Care Molecular Diagnostic Device. Sens Actuators B Chem 229, 232–238 (2016)
Y. Pan, T. Ma, Q. Meng, Y. Mao, K. Chu, Y. Men, T. Pan, B. Li, J. Chu, Droplet digital PCR enabled by microfluidic impact printing for absolute gene quantification. Talanta 211, 120680 (2020)
C.D. Ahrberg, B.R. Ilic, A. Manz, P. Neuzil, Handheld real-time PCR device. Lab. Chip 16(3), 586–592 (2016)
J.J. Chen, K.T. Li, Analysis of PCR kinetics inside a microfluidic DNA amplification system. Micromachines (Basel) 9(2) (2018)
V.L. Kopparthy, N.D. Crews, A versatile oscillating-flow microfluidic PCR system utilizing a thermal gradient for nucleic acid analysis. Biotechnol. Bioeng. 117(5), 1525–1532 (2020)
R.A. Mendoza-Gallegos, A. Rios, J.L. Garcia-Cordero, An affordable and portable thermocycler for real-time PCR made of 3D-Printed parts and off-the-Shelf Electronics. Anal. Chem. 90(9), 5563–5568 (2018)
Z. Li, R. Ju, S. Sekine, D. Zhang, S. Zhuang, Y. Yamaguchi, All-in-one microfluidic device for on-site diagnosis of pathogens based on an integrated continuous flow PCR and electrophoresis biochip. Lab. Chip 19(16), 2663–2668 (2019)
C. Shan, C. Zhang, J. Liang, Q. Yang, H. Bian, J. Yong, X. Hou, F. Chen, Femtosecond laser hybrid fabrication of a 3D microfluidic chip for PCR application. Opt. Express 28(18), 25716–25722 (2020)
N. Pak, D.C. Saunders, C.R. Phaneuf, C.R. Forest, Plug-and-play, infrared, laser-mediated PCR in a microfluidic chip. Biomed. Microdevices 14(2), 427–433 (2012)
B. Yang, P. Wang, Z. Li, C. Tao, Q. You, S. Sekine, S. Zhuang, D. Zhang, Y. Yamaguchi, A continuous flow PCR array microfluidic chip applied for simultaneous amplification of target genes of periodontal pathogens. Lab. Chip 22(4), 733–737 (2022)
C. Tzivelekis, M.P. Selby, A. Batet, H. Madadi, K. Dalgarno, Microfluidic chip fabrication and performance analysis of 3D printed material for use in microfluidic nucleic acid amplification applications. Journal of Micromechanics and Microengineering 31(3) (2021)
W. Cho, J.H. Maeng, Y. Ahn, S.Y. Hwang, Disposable on-chip microfluidic system for buccal cell lysis, DNA purification, and polymerase chain reaction. Electrophoresis 34(17), 2531–2537 (2013)
X. Zhu, J. Zhao, A. Hu, J. Pan, G. Deng, C. Hua, C. Zhu, Y. Liu, K. Yang, L. Zhu, A Novel Microfluidic device Integrated with Chitosan-Modified Capillaries for Rapid ZIKV Detection. Micromachines (Basel) 11(2) (2020b)
L. Xu, H. Qu, D.G. Alonso, Z. Yu, Y. Yu, Y. Shi, C. Hu, T. Zhu, N. Wu, F. Shen, Portable integrated digital PCR system for the point-of-care quantification of BK virus from urine samples. Biosens. Bioelectron. 175, 112908 (2021)
L. Gorgannezhad, K.R. Sreejith, J. Zhang, G. Kijanka, M. Christie, H. Stratton, N.T. Nguyen, Microfluidic array chip for parallel detection of waterborne Bacteria. Micromachines (Basel) 10(12) (2019)
S. Zhou, T. Gou, J. Hu, W. Wu, X. Ding, W. Fang, Z. Hu, Y. Mu, A highly integrated real-time digital PCR device for accurate DNA quantitative analysis. Biosens. Bioelectron. 128, 151–158 (2019)
J.J. Chen, S.J. Chen, Y.W. Ko, Continuous-Flow polymerase chain reaction chip by Water cooling. Eng. Agric. Environ. Food Chem 6(3), 99–104 (2013a)
J.J. Chen, C.M. Shen, Y.W. Ko, Analytical study of a microfludic DNA amplification chip using water cooling effect. Biomed. Microdevices 15(2), 261–278 (2013b)
Y. Alihosseini, M.R. Azaddel, S. Moslemi, M. Mohammadi, A. Pormohammad, M.Z. Targhi, M.M. Heyhat, Effect of liquid cooling on PCR performance with the parametric study of cross-section shapes of microchannels. Sci. Rep. 11(1), 16072 (2021)
A. Jalili, M. Bagheri, A. Shamloo, A.H. Kazemipour Ashkezari, A plasmonic gold nanofilm-based microfluidic chip for rapid and inexpensive droplet-based photonic PCR. Sci. Rep. 11(1), 23338 (2021)
T. Gou, J. Hu, W. Wu, X. Ding, S. Zhou, W. Fang, Y. Mu, Smartphone-based mobile digital PCR device for DNA quantitative analysis with high accuracy. Biosens. Bioelectron. 120, 144–152 (2018)
W. Liu, A. Warden, J. Sun, G. Shen, X. Ding, Simultaneous detection of multiple HPV DNA via bottom-well microfluidic chip within an infra-red PCR platform. Biomicrofluidics 12(2), 024109 (2018)
C.-J. Lee, Y.-H. Hsu, A size reduction method for rapid digital PCR using thin-film chip and vacuum pouch microfluidic system. Microfluidics and Nanofluidics 26(1) (2021)
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Kaixin sun, Yiqiang Fan, Yajun Zhang YumengXie, and KunMing Liang wrote the main manuscript text;
Ben Whiteside and Michael Hebda prepared Figs. 1, 2 and 3.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Competing interests
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
sun, K., Whiteside, B., Hebda, M. et al. A low-cost and hand-hold PCR microdevice based on water-cooling technology. Biomed Microdevices 25, 12 (2023). https://doi.org/10.1007/s10544-023-00652-4
Accepted:
Published:
DOI: https://doi.org/10.1007/s10544-023-00652-4