Abstract
Circulating tumor cells (CTCs) have been regarded as the major cause of metastasis, holding significant insights for tumor diagnosis and treatment. Although many efforts have been made to develop methods for CTC isolation and release in microfluidic system, it remains significant challenges to realize highly efficient isolation and gentle release of CTCs for further cellular and bio-molecular analyses. In this study, we demonstrate a novel method for CTC isolation and release using a simple wedge-shaped microfluidic chip embedding degradable znic oxide nanorods (ZnNRs) substrate. By integrating size-dependent filtration with degradable nanostructured substrate, the capture efficiencies over 87.5% were achieved for SKBR3, PC3, HepG2 and A549 cancer cells spiked in healthy blood sample with the flow rate of 100 μL min−1. By dissolving ZnNRs substrate with an extremely low concentration of phosphoric acid (12.5 mM), up to 85.6% of the captured SKBR3 cells were released after reverse injection with flow rate of 100 μL min−1 for 15 min, which exhibited around 73.6% cell viability within 1 h after release to around 93.9% after re-cultured for 3 days. It is conceivable that our microfluidic device has great potentials in carrying on cell-based biomedical studies and guiding individualized treatment in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Circulating tumor cells (CTCs) are rare cells escaped from primary tumor site and transported to distant organs through circulatory blood system, which is known as main cause of metastasis (Chambers et al. 2002; Fidler 2003). In the process of metastasis, these disseminated cells can be regarded as a “liquid biopsy” of the tumor, providing more vital biological and clinical insights for cancer diagnosis and treatment, such as determining tumor status, predicting therapeutic response, revealing the evolution of tumor cell heterogeneity. Over the past decade, several technologies have been utilized to meet the challenge of isolating and detecting CTCs from cancer patient blood samples. These approaches exploit different cell enrichment mechanisms, including immunomagnetic separation (Saliba et al. 2010; Luk et al. 2011; Jo et al. 2014, 2015; Esmaeilsabzali et al. 2016), microfluidics-based technologies (Nagrath et al. 2007; Stott et al. 2010; Ohnaga et al. 2013; Xue et al. 2015; Geislinger et al. 2015; Lee et al. 2017), centrifugal force (Hou et al. 2013a; Warkiani et al. 2016), size-based isolation (Tan et al. 2009; Hosokawa et al. 2010; Hosokawa et al. 2013; Tang et al. 2014; Sollier et al. 2014; Zhou et al. 2014), dielectrophoretic (DEP) separation (Guo et al. 2010; Jung et al. 2011; Huang et al. 2014b; Chou et al. 2017) and the enhanced interaction between nanostructured substrate and nanoscale components of the cellular surface (Zhang et al. 2012; Wang et al. 2017) .
Besides pursuing satisfied performance of CTC capture, how to realize gentle releasing and harvesting of captured CTCs with high viability for further culture and downstream molecular analysis is another critical issue, such as enzyme-linked immunesorbent assay (ELISA), polymerase chain reaction (PCR), enrichment and cultivation studies. For this purpose, several technologies have been employed to carry out on-chip CTC release and collection. Release of captured cells has been realized by a variety of approaches such as enzyme degradation (Adams et al. 2008; Shen et al. 2013; Yu et al. 2015), thermodynamic release (Hou et al. 2012; Liu et al. 2012a; Ke et al. 2015; Reátegui et al. 2015; Cheng et al. 2017), electrochemical desorption (Persson et al. 2011; Zhang et al. 2013; Jeon et al. 2014), laser irritation (Hou et al. 2013b), chemical reagent-triggered substrate sacrifice (Liu et al. 2013; Xie et al. 2014; Huang et al. 2014a, 2015) and light-responsive molecular conformation changes (Shin et al. 2014; Lv et al. 2015). However, majority of these methods have certain limitations such as low release efficiency, poor cell viability, and limited processing conditions. Further, these methods usually suffer from negative effects due to the imposed membrane stress, Joule heating, and thermal decomposition. Because high voltage and light irradiation with high energy can lead to unexpected damages to the recovered cells in a certain extent, which is unacceptable for subsequent cell culture and single cell analysis.
Herein, we present a novel method for highly efficient isolation and gently release of CTCs using a simple microfluidic chip which contained a wedge-shaped microchannel with gradually decreasing height and a layer of degradable znic oxide nanorods (ZnNRs) substrate. The ZnNRs were prepared by low-temperature thydrothermal method on clean glass slide (Guo et al. 2015). The morphology and crystal structure analysis of vertically aligned ZnNRs were investigated. And the microchannel was fabricated by wet etching technique and the whole microchip was assembled by thermal bonding technique. To demonstrate the enhanced cell-capture performance of microchannel with ZnNRs substrate comparing with pure glass substrate, a set of contrast experiments were performed for cell-capture testing. To investigate the effect of flow rate on capture efficiency for SKBR3 cells, a series of experiments were carried out with different flow rates (100, 200, 300, 400 and 500 μL min−1). To further evaluate the capture performance under optimized conditions (outlet height of 5 μm and flow rate of 100 μL min−1), artificial blood samples were utilized to perform cell capture assays by spiking SKBR3, PC3, HepG2, and A549 cells in healthy blood sample. A simple and friendly method was utilized to release the target cells by dissolving ZnNRs substrate with dilute phosphoric acid reversely introduced into the microchannel from outlet. To investigate how the flow rate affect release efficiency, a set of release experiments were carried out with different flow rates (50, 100, and 150 μL min−1). Also, the cell viabilities of SKBR3 and PC3 cells were investigated with different concentrations of phosphoric acid after releasing from microchip with ZnNRs substrate. The released SKBR3 cells and a certain number of leukocytes were collected and co-culture at 37 °C in 5% humidified CO2 incubator. After culture for 1, 2 and 3 days, the viability of SKBR3 cells were tested using FDA/PI staining.
2 Materials and methods
2.1 Materials and reagents
Anhydrous ethanol, acetone, zinc nitrate hexahydrate, hexamethylenetetramine, and phosphoric acid were obtained from Sinopharm Chemical Reagent Co., Ltd., China. Fluorescein diacetate (FDA), propidiumiodide (PI) and hoechst dye (no. 33342) were purchased from Sigma-Aldrich, USA. Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), and 1 × PBS (pH 7.4) were obtained from Life Technologies, USA. Trypsin (0.25%, w/v) was purchased from Gibco, USA. Penicillin/streptomycin solution was purchased from Genom, China. DI water was generated by a MILLI-Q system (Millipore, MA, USA). Blood samples were obtained from Zhongnan Hospital of Wuhan University, China.
2.2 Preparation and characterization of ZnNRs substrate
Glass slides were first washed by ethanol and dried by nitrogen stream. Then, zinc oxide (ZnO) thin film grown on the clean glass surface by using magnetron sputtering. Subsequently, thydrothermal method was performed to explore the growth of vertically aligned ZnNRs array on the exposed seed layer of ZnO nanocrystals in aqueous solution containing 25 mM zinc nitrate hydrate and 25 mM hexamethylenetetramine. The surface of glass slide with ZnO thin films was mounted toward the bottom of the reaction kettle placed in an oven and maintained at 95 °C for 6 h. Finally, the obtained glass slides with ZnNRs array were rinsed with DI water and dried in the oven at 80 °C for 30 min. The morphology of ZnNRs on glass substrate was investigated via Field Emission Scanning Electron Microscopy (S-4800, Hitachi, Janpan). X-ray diffraction (XRD) analysis was performed using X-ray diffractometer equipped with Cu-Kα radiation (D8GADDS, Bruker, Germany).
2.3 Fabrication of microfluidic chip
The microfluidic chip was fabricated by wet etching technique and thermal bonding technique. Figure 2a shows that the chip was composed of two glass slides. The top slide contained a wedge-shaped microchannel with gradually decreasing height from 60 μm to 7 μm, which was fabricated as followed procedures: 1) A layer of chemically corrosion-resistant adhesive tape was coated on standard glass slide (75 mm × 25 mm), the laser ablation system was utilized to transfer the microchannel design onto the adhesive tape. 2) The glass slide coated with patterned tape was immersed into glass etching solution (HF: HNO3: H2O = 10: 7: 83) at a low speed of around 0.65 mm min−1 to fabricate the microchannel with gradually decreasing height. 3) Two holes (0.5 mm in diameter) were drilled by laser on the top glass slide as the inlet and outlet of the microchannel, respectively. The bottom slide contained vertically aligned ZnNRs (around 2 μm height) array on the surface of the glass substrate with the same pattern as upper microchannel. Finally, the top and bottom glass slides were bonded together after a dynamic heating and annealing process in a programmable muffle furnace.
2.4 Cell culture
SKBR3 (human breast cancer cell), PC3 (human prostate cancer cell), HepG2 (human liver cancer cell) and A549 (human lung cancer cell), were obtained from China Center for Type Culture Collection (CCTCC). Cells were cultured in Dulbecco’s modified eagle medium (DMEM) added with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in 5% humidified CO2 incubator. Adherent cells were released with 0.25% (w/v) trypsin, resuspended in DMEM media, and rocked gently on a shaker 5 min prior to experiments.
2.5 Cell capture and release
Before the experiment, cells were prestained with Hochest 33,342 (10 μg mL−1) for 10 min. Stained cells were spiked in DMEM solution at a concentration of 200 cells mL−1. In order to investigate how flow rate affected the capture efficiency, the spiked samples (SKBR3) were injected into microchips with the outlet height of 5 μm at the flow rates of 100, 200, 300, 400, and 500 μL min−1, respectively, followed by rinsing with PBS at flow rate of 50 μL min−1 for 5 min to continue pumping remained cells in the perfusion tube. To evaluate the capture performance of the microchip, four kinds of artificial blood samples were prepared by spiking prestained SKBR3, PC3, HepG2, and A549 cells in healthy blood sample at a concentration of 200 cells mL−1, respectively. And then the spiked samples were injected into microchips at the flow rate of 100 ml−1. After being rinsed with PBS, the cells escaped from the microchips were collected in a vial. Cells remaining in the microchip and in the collection vial were both imaged and counted manually using a fluorescence microscope. To achieve cell release, dilute phosphoric acid solution with optimized concentration (12.5 mM) was introduced into the microchannel from the outlet of microchip with the flow rate of 50, 100 and 150 μL min−1 for 15 min, respectively. As the degradation of ZnNRs substrate, captured cells escaped from the microchannel under reverse hydrodynamic force. Finally, the remained and released cells were both imaged and counted for calculating release efficiencies.
2.6 Viability assay of released cancer cell
To test the cell viability after cell release, a fluorescent live-dead staining assay was utilized to visualize the proportion of viable cells after release. To obtain a high viability of the released cells, optimization assays were performed to investigate how the concentration of dilute phosphoric acid solution affected the viabilities of released SKBR3 and PC3 cells at flow rate of 100 μL min−1 for 15 min. The harvested solution containing the released cells was centrifuged (1500 r min−1, 2 min) and washed with PBS for 3 times to remove residual solution. Then, the harvested cells were resuspended in staining solution containing FDA (10 μg mL−1) and PI (2 μg mL−1) and cultivated at 37 °С in 5% humidified CO2 incubator for 10 min. After washing with PBS for 2 times, a drop of cell suspension was placed on the culture dish and covered by a coverslip for following observation. Imaging and counting of the live/dead cells were performed with a CCD camera (DP72, Olympus, Japan) mounted on an inverted fluorescence microscope (IX81, Olympus, Japan).
2.7 Re-culture of the released cells
In re-culture experiment, SKBR3 cells spiked in healthy blood sample without perstaining treatment were applied to perform cell capture and release under optimized conditions. After collection and washing with PBS for 3 times by low-speed centrifugation, the harvested cells were resuspended in DMEM and transferred into a 48-well plate and cultivated at 37 °C under a humidified 5% CO2 atmosphere for 1, 2 and 3 days. The culture medium was replaced once a day. FDA/PI staining was implemented to determine the viability of re-cultured cells. In addition, sterile conditions should be maintained during the whole procedure.
3 Results and discussions
3.1 Design of the microfluidic chip
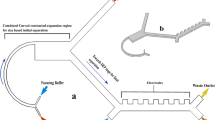
As shown in Fig. 1, our cancer cell capture/release microchip contained a wedge-shape microchannel with gradually decreasing height of 60 μm to 5 μm and embedded a layer of degradable ZnNRs (around 2 μm) substrate. The wedge-shaped microchannel was designed to capture and locate the cancer cells based on the biophysical property that CTCs are stiffer, larger sizes, and larger nuclear/cytoplasmic ratios compared with white/red blood cells. Meanwhile, the frictional resistance between cellular surface components of cancer cells and nanostructured substrate was enhanced comparing with red/white blood cells which have non-rough surface, resulting in much higher capture efficiency for cancer cells. Utilizing the special design, the microfluidic chip could efficiently isolate smaller CTCs with a very small cut-off size from patient blood sample without clogging. Due to the rapid and efficient isolation, our method can realize preliminary screening of CTCs from large volume of blood sample (up to 10 mL), and remove the white blood cells as far as possible (with removal efficiency of 99.95%) under the prerequisite of high capture efficiency (more than 85%) of CTCs. After that, more sensitive CTC identified methods such as FISH testing, IF staining, PCR assay and gene sequencing could take its time to be performed. Due to the easy degradablility of ZnNRs substrate, the captured cells can be efficiently released and collected from the inlet of microfluidic chip with high viability when dilute phosphoric acid solution was introduced into the microchannel from the outlet with a low flow rate, and the damages to the released cells caused by hydrodynamic force are almost negligible.
3.2 Characterization of ZnNRs array
Low-temperature hydrothermal method was utilized to grow ZnNRs on the seed layer of ZnO nanocrystals on clean glass slide. As shown in Fig. 2b, the layer of ZnNRs array was relatively homogeneously grown on the bottom glass slide of the microchannel. The SEM images of ZnNRs array were shown in Fig. 2c (top view) and Fig. 2d (cross sectional view). The magnified SEM image of the vertically aligned ZnNRs showed quasi-hexagnal morphology, and the ZnNRs ranged from 150 to 300 nm in diameter and up to around 2 μm in length. To confirm the composition of the thin layer, XRD measurement was carried out. Figure 2e showed the wide range (25° to 70°) XRD 2θ-scan of ZnNRs array. Only one diffractive peak at 34.5° was observed, which corresponds to the (002) direction of the ZnO diffractive peak. This result indicates that the ZnNRs are in the c-axis preferred orientation.
a Design of the CTC capture/release microchip contained a wedge-shape microchannel with gradually decreasing height of 60 μm to 5 μm and embedded a layer of degradable ZnNRs (around 2 μm) substrate. b The digital photograph of microfluidic chip. c-d The top and cross-section SEM images of the vertically aligned ZnNRs array grown on glass substrate. Scale bars, 1 μm. e Wide-range XRD 2θ-scan of the ZnNRs array
3.3 Cell-capture performance testing
To demonstrate the enhanced cell-capture performance of microchannel with ZnNRs substrate comparing with pure glass substrate, two different kinds of microfluidic chips (with/without ZnNRs substrate) were fabricated and applied for cell-capture testing. To investigate how the flow rate affected the capture efficiencies for SKBR3 cells spiked in DMEM, the spiked samples were injected into microfluidic devices with a fixed outlet height of 5 μm with different flow rates (100, 200, 300, 400, and 500 μL min−1). The capture efficiency was defined as the ratio of the number of captured target cells to the number initially introduced. As shown in Fig. 3a, the cell-capture efficiency of microchip with ZnNRs substrate decreased more slowly with increasing flow rate from 100 to 500 μL min−1 compared with pure glass substrate. It suggested that the enhanced capture efficiency benefitted from the frictional resistance between cellular surface components (e.g., microvilli and filopodia) and the nanostructured substrate. As a result, flow rate of 100 μL min−1 was chosen as the ideal injecting parameter which guaranteed at least 90% capture efficiencies for SKBR3 cells.
a Capture efficiency of spiked SKBR3 cells at different flow rate in two kinds of microfluidic chips with/without ZnNRs substrate. b Capture efficiencies of spiked four different cancer cells (i.e., SKBR3, PC3, HepG2 and A549) in healthy blood sample with the flow rate of 100 μL min−1 in these two kinds of microfluidic chips
To further test the cell-capture performance under the optimal capture condition (100 μL min−1 of flow rate), four kinds of artificial blood samples were prepared by spiking perstained SKBR3, PC3, HepG2, and A549 cells in healthy blood sample and then introduced into microchips. As shown in Fig. 3b, the average capture efficiencies of more than 85% and 80% were achieved for above four artificial blood samples in microchips with/without ZnNRs substrate, respectively.
3.4 Cell-release and viability assay
To achieve the full availability of CTCs for further cellular and molecular analysis, it is necessary to gently release cancer cells and ensure the viability. Herein, a simple and friendly method was utilized to release the target cells by dissolving ZnNRs substrate with dilute phosphoric acid reversely introduced into the microchannel from outlet. To obtain a good balance between release efficiency and cell viability, the concentration of 12.5 mM and the rinsing time of 15 min were chosen as the optimal releasing conditions. To investigate the influence of flow rate on the release efficiency, a set of release experiments were performed under different flow rates (50, 100, and 150 μL min−1). ZnNRs substrate (around 2 μm in thickness) can be dissolved with low concentration of phosphoric acid solution at room temperature.
ZnO was reduced by phosphoric acid and water-soluble Zn2+ ions were released into the solution (Eq. 1). The ZnNRs substrate was dissolved and the height of the microchannel was increased by around 2 μm, resulting in the release of the target cells were from the original captured sites. As shown in Fig. 4a, the release efficiency increased with the increasing of flow rate for microchip with/without ZnNRs substrate. An average of 85.6% release efficiency was obtained for microchips with ZnNRs substrate at the flow rate of 100 μL min−1, but only 53.9% release efficiency for microchips with pure glass substrate. Figure 4b showed fluorescent images of the captured cells on ZnNRs and pure glass substrate before and after release. With the approach of scarification of ZnNRs substrate, the captured cells could be rapidly and efficiently released with low damage, which has great potential for further biochemical and biomedical analysis.
a The relationship of the release efficiencies of spiked SKBR3 cells and the flow rate in microfluidic chips with/without ZnNRs substrate with the reverse injection of phosphoric acid (12.5 mM) for 15 min. b Fluorescent images of SKBR3 cells before and after release in microchips with/without ZnNRs substrate. Scale bars, 100 μm
Benefiting to the specific designed microchip, CTCs would be rapid and efficiently isolated based on the biophysical property and the frictional resistance. Then further recognization of CTCs could be performed such as FISH testing, IF staining, PCR assay and gene sequencing. After that, the identified CTCs are able to be further purified by utilizing cell sorting microfluidic devices (Guo et al. 2010; Liu et al. 2012b).
A series of experiments was performed on microchips with ZnNRs substrate to further investigate the dependence of cell release viability on the concentration of phosphoric acid. Captured SKBR3 and PC3 cells were released and collected with different concentrations of phosphoric acid (0, 12.5, 25, 50, and 75 mM). As shown in Fig. 5a, the average viabilities were 73.6% for released SKBR3 cells and 68.9% for released PC3 cells when the concentration was lower than 12.5 mM, but the average viabilities dropped down to 9.2% for SKBR3 cells and 5.6% for PC3 cells when the concentration was 25 mM.
a Cell viabilities of released SKBR3 and PC3 cells under different concentrations of phosphoric acid at room temperature. b Cell viabilities of released SKBR3 cells after centrifugation within 1 h and re-cultured for 1, 2 and 3 days. c Bright field and fluorescent images of SKBR3 cells re-cultured for 1, 2 and 3 days, respectively. Scale bars, 100 μm
3.5 Re-culture of release cells
For further test the availability of the re-culture of the harvested CTCs, the released cancer cells and a certain number of leukocytes were collected and co-culture in the same cell culture flask. After exchanging the culture medium once a day, the dead cells and leukocytes were removed and the viability of remained cells were tested using FDA/PI staining. As shown in Fig. 5b, an average viability increased from around 73.6% to 82.1% after re-cultured for 1 day, and the final viability can reach up to 93.9% after re-cultured for 3 days. The corresponding bright field images and fluorescence of re-cultured SKBR3 cells were shown in Fig. 5c. The results indicated that released cancer cells could be further expanded in culture and subsequent molecular and biomedical analyses.
4 Conclusions
In summary, a novel method for highly efficient isolation and gently release was demonstrated using a simple wedge-shaped microfluidic device embedding degradable ZnNRs substrate. By integrating size-dependent filtration with degradable nanostructured substrate, the capture efficiencies over 85% were achieved for SKBR3, PC3, HepG2 and A549 cancer cells spiked in healthy blood sample at the flow rate of 100 μL min−1. Due to the frictional resistance between cellular surface components (e.g., microvilli and filopodia) and the nanostructured substrate, the capture efficiency of microchips with ZnNRs substrate was enhanced comparing to microchips with pure glass substrate. Besides, the microfluidic chip can enrich CTCs without losing the small CTCs and avoiding the clogging issue owing to easier deformability of WBCs and disk-like RBCs than target cancer cells. A simple and friendly substrate-sacrificed method was performed by chemically etching ZnNRs substrate with dilute phosphoric acid (12.5 mM) to realize effectively releasing around 85.6% of captured SKBR3 cells with the flow rate of 100 μL min−1 for 15 min, which exhibited increasing cell viability from 73.6% after re-cultured for 1 day to 93.9% after re-cultured for 3 days. It is conceivable that the presented strategy may have potential applications in guiding individualized treatment, capturing and releasing viable CTCs for ex vivo culture, monitoring progression and estimating prognosis of malignant tumors.
References
A.A. Adams, P.I. Okagbare, J. Feng, M.L. Hupert, D. Patterson, J. Göttert, R.L. McCarley, D. Nikitopoulos, M.C. Murphy, S.A. Soper, J. Am. Chem. Soc. 130, 8633 (2008)
A.F. Chambers, A.C. Groom, I.C. MacDonald, Nat. Rev. Cancer 2, 563 (2002)
W.P. Chou, H.M. Wang, J.H. Chang, T.K. Chiu, C.H. Hsieh, C.J. Liao, M.H. Wu, Sensor. Actuat. B: Chem. 241, 245 (2017)
S.B. Cheng, M. Xie, Y. Chen, J. Xiong, Y. Liu, Z. Chen, S. Guo, Y. Shu, M. Wang, B.F. Yuan, W.G. Dong, W.H. Huang, Anal. Chem. 89, 7924 (2017)
H. Esmaeilsabzali, T.V. Beischlag, M.E. Cox, N. Dechev, A.M. Parameswaran, E.J. Park, Biomed. Microdevices 18, 22 (2016)
I.J. Fidler, Nat. Rev. Cancer 3, 453 (2003)
T.M. Geislinger, M.E.M. Stamp, A. Wixforth, T. Franke, Appl. Phys. Lett. 107, 203702 (2015)
F. Guo, X.H. Ji, K. Liu, R.X. He, L.B. Zhao, Z.X. Guo, W. Liu, S.S. Guo, X.Z. Zhao, Appl. Phys. Lett. 96, 193701 (2010)
Z. Guo, H. Li, L.Q. Zhou, D.X. Zhao, Y.H. Wu, Z.Q. Zhang, W. Zhang, C.Y. Li, J. Yao, Small 11, 438 (2015)
M. Hosokawa, T. Hayata, Y. Fukuda, A. Arakaki, T. Yoshino, T. Tanaka, T. Matsunaga, Anal. Chem. 82, 6629 (2010)
M. Hosokawa, H. Kenmotsu, Y. Koh, T. Yoshino, T. Yoshikawa, T. Naito, T. Takahashi, H. Hurakami, Y. Nakamura, A. Tsuya, T. Shukuya, A. Ono, H. Akamatsu, R. Watanabe, S. Ono, K. Mori, H. Kanbara, K. Yamaguchi, T. Tanaka, T. Matsunaga, N. Yamanoto, PLoS One 8, e67466 (2013)
S. Hou, H. Zhao, L. Zhao, Q. Shen, K.S. Wei, D.Y. Suh, A. Nakao, M.A. Garcia, M. Song, T. Lee, B. Xiong, S.C. Luo, H.R. Tseng, H.H. Yu, Adv. Mater. 25, 1514 (2012)
S. Hou, L. Zhao, Q. Shen, J. Yu, C. Ng, X. Kong, D. Wu, M. Song, X. Shi, X. Xu, Angew. Chem. Int. Ed. 52, 3379 (2013b)
H.W. Hou, M.E. Warkiani, B.L. Khoo, Z.R. Li, R.A. Soo, D.S.W. Tan, W.T. Lim, J. Han, A.A.S. Bhagat, C.T. Lim, Sci Rep 3, 1259 (2013a)
Q. Huang, B. Chen, R. He, Z. He, B. Cai, J. Xu, W. Qian, H.L. Chan, W. Liu, S. Guo, X.Z. Zhao, J. Yuan, Adv. Healthcare. Mater. 3, 1420 (2014a)
C. Huang, C. Liu, J. Loo, T. Stakenborg, L. Lagae, Appl. Phys. Lett. 104, 013703 (2014b)
C. Huang, G. Yang, Q. Ha, J. Meng, S. Wang, Adv. Mater. 27, 310 (2015)
S. Jeon, J.M. Moon, E.S. Lee, Y.H. Kim, Y. Cho, Angew. Chem. Int. Ed. 53, 4597 (2014)
S.M. Jo, S.H. Noh, Z. Jin, Y. Lim, J. Cheon, H.S. Kim, Sensor. Actuat. B: Chem. 201, 144 (2014)
S.M. Jo, J.J. Lee, W. Heu, H.S. Kim, Small 11, 1975 (2015)
J. Jung, S.K. Seo, Y.D. Joo, K.H. Han, Sensor. Actuat. B: Chem. 157, 314 (2011)
Z. Ke, M. Lin, J.F. Chen, J.S. Choi, Y. Zhang, A. Fong, A.J. Liang, S.F. Chen, Q. Li, W. Fang, P. Zhang, M.A. Garcia, T. Lee, M. Song, H.A. Lin, H. Zhao, S.C. Luo, S. Hou, H.H. Yu, H.R. Tseng, ACS Nano 9, 62 (2015)
T.Y. Lee, K.A. Hyun, S.I. Kim, H.I. Jung, Sensor. Actuat. B: Chem. 238, 1144 (2017)
H. Liu, X. Liu, J. Meng, P. Zhang, G. Yang, B. Su, K. Sun, L. Chen, D. Han, S. Wang, Adv. Mater. 25, 922 (2012a)
H. Liu, Y. Li, K. Sun, J. Fan, P. Zhang, J. Meng, S. Wang, L. Jiang, J. Am. Chem. Soc. 135, 7603 (2013)
K. Liu, Y.L. Deng, N.G. Zhang, S.Z. Li, H.J. Ding, F. Guo, W. Liu, S.S. Guo, X.Z. Zhao, Microfluid. Nanofluid. 13, 761 (2012b)
L.Y. Luk, C.M.L. Chan, A.H.K. Cheung, V.H.M. Lee, P.B.S. Lai, B.B.Y. Ma, E.P. Hui, M.Y.Y. Lam, T.C.C. Au, A.T.C. Chan, Br. J. Cancer 104, 1000 (2011)
S.W. Lv, J. Wang, M. Xie, N.N. Lu, Z. Li, X.W. Yan, S.L. Cai, P.A. Zhang, W.G. Dong, W.H. Huang, Chem. Sci. 6, 6432 (2015)
S. Nagrath, L.V. Sequist, S. Maheswaran, D.W. Bell, D. Irimia, L. Ulkus, M.R. Smith, E.L. Kwak, S. Digumarthy, A. Muzikansky, P. Ryan, U.J. Balis, R.G. Tompkins, D.A. Haber, M. Toner, Nature 450, 1235 (2007)
T. Ohnaga, Y. Shimada, M. Moriyama, H. Kishi, T. Obata, K. Takata, T. Okumura, T. Nagata, A. Muraguchi, K. Tsukada, Biomed. Microdevices 15, 611 (2013)
K.M. Persson, R. Karlsson, K. Svennersten, S. L€offler, E.W.H. Jager, A. Richter-Dahlfors, P. Konradsson, M. Berggren, Adv. Mater. 23, 4403 (2011)
E. Reátegui, N. Aceto, E.J. Lim, J.P. Sullivan, A.E. Jensen, M. Zeinali, J.M. Martel, A.J. Aranyosi, W. Li, S. Castleberry, A. Bardia, L.V. Sequist, D.A. Haber, S. Maheswaran, P.T. Hammond, M. Toner, S.L. Stott, Adv. Mater. 27, 1593 (2015)
A.E. Saliba, L. Saias, E. Psychari, N. Minc, D. Simon, F.C. Bidard, C. Mathiot, J.Y. Pierga, V. Fraisier, J. Salamero, V. Saada, F. Farace, P. Vielh, L. Malaquin, J.L. Viovy, Proc. Natl. Acad. Sci. U. S. A. 107, 14524 (2010)
Q. Shen, L. Xu, L. Zhao, D. Wu, W. Fan, Y. Zhou, W.H. OuYang, X. Xu, Z. Zhang, M. Song, T. Lee, M.A. Garcia, B. Xiong, S. Hou, H.R. Tseng, X. Fang, Adv. Mater. 25, 2368 (2013)
D.S. Shin, J. You, A. Rahimian, T. Vu, C. Siltanen, A. Ehsanipour, G. Stbayeva, J. Sutcliffe, A. Revzin, Angew. Chem. Int. Ed. 53, 8221 (2014)
E. Sollier, D.E. Go, J. Che, D.R. Gossett, S. O'Byrne, W.M. Weaver, N. Kummer, M. Rettig, J. Goldman, N. Nickols, S. McCloskey, R.P. Kulkarni, D.D. Carlo, Lab Chip 14, 63(2014)
S.L. Stott, C.H. Hsu, D.I. Tsukrov, M. Yu, D.T. Miyamoto, B.A. Waltman, S.M. Rothenberg, A.M. Shah, M.E. Smas, G.K. Korir, F.P. Floyd, A.J. Gilman, J.B. Lord, D. Winokur, S. Springer, D. Irimia, S. Nagrath, L.V. Sequist, R.J. Lee, K.J. Isselbacher, S. Maheswaran, D.A. Haber, M. Toner, Proc. Natl. Acad. Sci. U. S. A. 107, 18392 (2010)
S.J. Tan, L. Yobas, G.Y.H. Lee, C.N. Ong, C.T. Lim, Biomed. Microdevices 11, 883 (2009)
Y. Tang, J. Shi, S. Li, L. Wang, Y.E. Cayre, Y. Chen, Sci Rep 4, 6052 (2014)
W. Wang, H. Cui, P. Zhang, J. Meng, F. Zhang, S. Wang, ACS Appl. Mater. Interfaces 9, 10537 (2017)
M.E. Warkiani, B.L. Khoo, L. Wu, A.K.P. Tay, A.A.S. Bhagat, J. Han, C.T. Lim, Nat. Protoc. 11, 134 (2016)
M. Xie, N.N. Lu, S.B. Cheng, X.Y. Wang, M. Wang, S. Guo, C.Y. Wen, J. Hu, D.W. Pang, W.H. Huang, Anal. Chem. 86, 4618 (2014)
P. Xue, Y. Wu, J. Guo, Y. Kang, Biomed. Microdevices 17, 39 (2015)
X.L. Yu, B.R. Wang, N.G. Zhang, C.Q. Yin, H. Chen, L.L. Zhang, B. Cai, Z.B. He, L. Rao, W. Liu, F.B. Wang, S.S. Guo, X.Z. Zhao, Acs Appl. Mater. Inter. 7, 24001 (2015)
P.C. Zhang, T.L.X. Li Chen, H.L. Liu, X.L. Liu, J.X. Meng, G. Yang, L. Jiang, S.T. Wang, Adv. Mater. 25, 3566 (2013)
N.G. Zhang, Y.L. Deng, Q.D. Tai, L.Y. Hong, Q.L. Shen, B.R. Cheng, B. Xiong, H.-R. Tseng, S.S. Guo, K. Liu, X.Z. Zhao, Adv. Mater. 24, 2756 (2012)
M.D. Zhou, S. Hao, A.J. Williams, R.A. Harouaka, B. Schrand, S. Rawal, Z. Ao, R. Brennaman, E. Gilboa, B. Lu, S.W. Wang, J.Y. Zhu, R. Datar, R. Cote, Y.C. Tai, S.Y. Zheng, Sci Rep 4, 7392 (2014)
Acknowledgements
This work was supported in part by following foundations: (1) National Natural Science Foundation of China (81372358, 81527801, 51303140 and 81602489); (2) Natural Science Foundation of Hubei Province, China (2014CFA029); (3) Colleges of Hubei Province Outstanding Youth Science and Technology Innovation Team (T201305); (4) Applied Foundational Research Program of Wuhan Municipal Science and Technology Bureau (2015060101010056); (5) The Technology Research and Development Fund in Shenzhen (JCYJ20150403091443300, JCYJ20150403091443271); (6) The Natural Science Foundation of Guangdong (2016A030310242, 2016A030313381). We appreciate Professor Bin Xiong from Zhongnan Hospital of Wuhan University for providing breast cancer patient and healthy blood samples.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Nangang Zhang and Kan Liu are co-corresponding authors.
Electronic supplementary material
ESM 1
(DOCX 417 kb)
Rights and permissions
About this article
Cite this article
Li, S., Gao, Y., Chen, X. et al. Highly efficient isolation and release of circulating tumor cells based on size-dependent filtration and degradable ZnO nanorods substrate in a wedge-shaped microfluidic chip. Biomed Microdevices 19, 93 (2017). https://doi.org/10.1007/s10544-017-0235-7
Published:
DOI: https://doi.org/10.1007/s10544-017-0235-7