Abstract
Much of the recent philosophical debate on causation and causal explanation in the biological and biomedical sciences has focused on the notion of mechanism. Mechanisms, their nature and epistemic roles have been tackled by a range of so-called neo-mechanistic theories, and widely discussed. Without denying the merits of this approach, our paper aims to show how lately it has failed to give proper credit to processes, which are central to the field, especially of contemporary molecular biology. Processes can be summed up in the notion of ‘pathway’, which is far from being just equivalent to that of ‘mechanism’ and has a profound epistemological and explanatory relevance. It is argued that an adequate consideration of pathways impels some rethinking of scientific explanation in molecular biology, namely its functional and contextual features. A number of examples are given to suggest that the focus of philosophical attention in this disciplinary field should shift from the notion of mechanism to the notion of pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Mechanisms and the new mechanical philosophy: What’s new?
As is widely known, the philosophical position of mechanism has held sway in the last few decades. Ever since Wesley Salmon’s probabilistic mechanism, the notion of mechanism has been advocated as central to the debate on causation and causal explanation. A plethora of theories have been put forward to provide the most appropriate definition for a number of specific disciplinary fields, or possibly all of them. Mechanisms have thus been discussed with respect to, for instance, economics, history, psychology, and, especially, biology. In parallel with attempts to revise and refine such a notion to adapt it to specific disciplinary fields, and related epistemic purposes, a growing trend in the literature has sought to devise a one-fits-all definition to cover roughly any case. That is how we have ended up with some of the most recent accounts of what is meant by ‘mechanism’, such as: “A mechanism for a phenomenon consists of entities and activities organized in such a way that they are responsible for the phenomenon” (Illari and Williamson 2012, p. 120), or: “a mechanism for a phenomenon consists of entities (or parts) whose activities and interactions are organized so as to be responsible for the phenomenon” (Glennan 2017, p. 17).
The literature on mechanisms in contemporary philosophy of science is so vast it cannot be exhaustively tackled here. Suffice to say that the latest contributions, basically from the late Nineties onwards, now fall under the umbrella of the so-called “new mechanical philosophy”.Footnote 1 Although neo-mechanistic theories differ from each other, the overall purpose of this philosophical trend is to have some notion which can work for “mechanisms across the sciences” (Illari and Williamson 2012), and which is “grounded in the details of scientific practice” (Machamer et al. 2000). While having many merits in these respects, neo-mechanistic theories have also been subject to criticism and to attempts at further clarification (see e.g. Levy 2013) to disentangle aspects which risk being conflated. While Salmon’s main concern back in the Eighties was to provide strict criteria to tell what counted as genuinely causal from what did not, more recent positions aim to account for a wide range of relations and systems deemed “causal”. Neo-mechanists thus dwell on different kinds of activities and entities, different types of mechanisms, and a variety of causal claims (see e.g. Glennan 2017, chs. 4–6; Glennan and Illari 2017a, b). “Mechanistic diversity” is at stake: instead of sharp distinctions between mechanisms and non-mechanisms, we seem to be facing a sort of continuum, along which different cases can be more or less paradigmatic of a mechanism—or, rather, marginal—according to a set of parameters (see Skillings 2015).Footnote 2
If diversity reigns, vagueness lies in ambush: given their breadth, definitions such as those quoted above—or even previous ones, like Machamer et al. (2000)—might be hard to disagree with, but equally hard to promote as thoroughly fruitful. Generality and the widest applicability combined with close adherence to actual scientific practice might be goals difficult to achieve at the same time. The “strategic vagueness” regarding their application might “limit the vulnerability” of many neo-mechanist claims (Rosenberg 2018, p. 11), but at the price of making them methodologically unproductive, at least for some scientific fields. Among warnings on possible ambiguities or shortcomings, we shall here focus on some raised by John Dupré (2013) and by Stavros Ioannidis and Stathis Psillos (2017), which we regard as particularly relevant to our proposals and arguments in what follows. We believe they can help in re-discussing the adequacy of neo-mechanism with respect to scientific enquiries, and avoid the looming risk of some “anything-counts-as-a-mechanism” view.
According to Dupré, recent philosophical theories of causation fail to provide a satisfactory account of the complexity of biological phenomena, insofar as they do not devote serious attention to the “thoroughly processual character of living systems” (Dupré 2013, p. 19). Starting from a critical reading of Jon Williamson’s account of epistemic causality, its purposes and feasibility, Dupré puts forward a rather radical processual account of living beings—which, he claims, should differ from extant process-based accounts of causality. The latter prove to be wholly related to strictly physical causes, and hence hardly translatable into biological contexts and unable to convey the complex relation between mechanisms and functions. Given the broad use of the term “mechanism” in biology—an aspect that neo-mechanists are generally keen to stress—the notion of mechanism can ultimately be philosophically misleading: “there is a serious danger of vacuity in some treatments [of biological systems], in which it seems that mechanisms just are whatever explains whatever happens” (ibid., p. 28). We must not only be able to tell what does not count as a mechanism, as Dupré himself points out, but also, in a constructive spirit, wonder whether some other notion can better accommodate explanations of biological phenomena. Instead of thinking of biological systems in terms of neo-mechanist accounts, with their inventories of entities and interactions, we would better be aware that “what are stable and robust in biology are not things, but processes” (ibidem, italics added. See also Nicholson and Dupré, forthcoming).
For their part, Ioannides and Psillos advocate methodological mechanism, taken as the search for causal pathways and incorporating “a minimal account of understanding mechanisms”. Picking apoptosis as a case study, they argue that “a mechanism just is a causal pathway described in the language of theory” (Ioannides and Psillos 2017, p. 601, italics added).Footnote 3 Their deflationary account of mechanisms is meant to differ from current accounts in that it does not imply any ontological commitment, and strongly warns against attempts to draw metaphysical conclusions from premises extracted from scientific practice.Footnote 4 In scientific practice—they claim—scientists’ talking about mechanisms actually refers to the search for a causal pathway, which is taken to explain how the phenomenon under investigation is brought about. Efforts are driven towards the identification of the causal pathway, not towards some understanding of specific theories of causation that might account for it, or towards its characterization in terms of entities, activities, interactions, powers, and the like. It is worth stressing that the hallmark of mechanistic explanations is their explaining not only what brought a given effect about, but how it did so, and this role is here attributed to causal pathways. Ioannidis and Psillos’ view questions both extant mechanistic theories, and, more generally, the drawing of theoretical conclusions from the use of certain notions in scientific practice.
While motivated by different philosophical concerns, the proposals presented by the three authors just mentioned can be seen to intersect insofar as they show a renewed interest in a processual conception of causation. Whereas Salmon (1984, 1989, 1998) and later Phil Dowe (2000)Footnote 5 suggested that causation was to be thought of in terms of interacting processes, neo-mechanism has opted for an entity ontology. In principle, this would provide a much more comprehensive and so-to-speak flexible ontology, with bottoming-out strategies being directly oriented by the specific discipline at stake in the investigation pursued. Has the shift away from processes been entirely advantageous, or at least has it ensured more benefits than downsides in all fields and respects? What follows questions whether this has actually been the case by taking a close look at some scientific literature in molecular biology, the scientific field our analysis addresses. We will argue that the notion of pathway plays a role hitherto underestimated by the new mechanical philosophy, and conclude that it might help us rethink the very features of explanatory strategies in molecular biology. Our focus will not be on metaphysical issues—which Dupré touches upon—but—more in line with the spirit of Ioannides and Psillos’ work—on strictly methodological problems. Unlike Ioannides and Psillos, however, we shall stress a few differences between the notions of pathway and mechanism, and possible implications that such differences might have with respect to explanation. We maintain that a very close and careful analysis of actual cases in scientific practice, such as those considered below, shows the necessary role of the notion of pathway and fosters a processual approach to explanatory issues in molecular biology. While not strictly requiring a commitment to a processual metaphysics, our claims are by no means incompatible with such a framework, but can rather be taken as providing some support. However, this argument would also require—we believe—further considerations on which metaphysical conclusions are to be drawn from scientific practice, and this falls beyond the specific scope of this paper.Footnote 6
Functional explanation, molecular pathways and their contexts
From mechanisms to pathways
The revival of mechanisms in contemporary philosophical literature, back in the Eighties, stemmed from the search for some sound notion of causation that might help tell genuine causes from mere statistical correlations. As recalled above, many efforts were then devoted to devising criteria that could distinguish genuine causal processes from pseudoprocesses—criteria that sharpened concepts that ultimately proved unable to do much philosophical work (see Hitchcock 2004). Ever since, mechanistic philosophy has been struggling to make sense of some notion of causation that could account for the myriad cases of production sciences are interested in, with an increasing consideration of the work of biologists, especially in such fields as molecular biology, neurobiology, biochemistry and cell biology (see e.g. Darden 2008; Craver and Darden 2013). Do present accounts of mechanistic causation manage to capture what biologists themselves provide as explanations and what they are looking for? How can we make sure that the notion of mechanism, far from being just some sort of filler term or providing illusionary steps forward in our understanding, actually enables us to grasp (at least some features of) scientific investigations on the “causal structure of the world” (see Salmon 1984)? Neo-mechanists have largely stated that they are concerned with the sciences as they are actually practised: when addressing mechanistic explanation, they should thus not explain “how things work” in the natural realm, but rather “how things work” when scientists themselves show “how things work” (Franklin-Hall 2016, p. 70). Laura Franklin-Hall has recently argued that, if, on the one hand, mechanists have been right in claiming that the received view of explanation was too far from actual science to shed light on it, on the other hand, they have remained too close to science itself to say anything interesting about it. Is this really the case? Have they in actual fact remained too close to science, and can all mechanist and neo-mechanist hints be taken on a par?
As recalled in “Mechanisms and the new mechanical philosophy: What’s new?” section, Ioannides and Psillos define mechanisms in terms of pathways, for methodological purposes. While sympathising with their appeal to the notion of pathway, and sharing their purely methodological standpoint, we argue that a close look at the scientific literature in molecular biology might question the conceptual equivalence between mechanisms and pathways, and thereby stimulate further philosophical considerations on what explanation amounts to in this disciplinary context.
From a close look at current molecular biology literature, a couple of things spring to mind. In the first place, there is a pervasive use of the word “mechanism”, but a further look reveals the term is used loosely, usually in an allusive rather than precise sense, and often evocatively. What is worth stressing is scientific papers provide no clear, univocal and sharp definition of what “mechanism” actually stands for. Secondly, molecular biology makes even use of the notions of mechanism, process and pathway—with processes and pathways being basically regarded as equivalent in this context. If, then, philosophical interest in mechanisms is taken to be motivated also by the number of occurrences of ‘mechanism’ in molecular biology (Bechtel and Abrahamsen 2005, pp. 422–423), the same argument should apply to support philosophical inquiries into the notion of pathway. One advantage in dealing with a pathway is that, unlike the notion of mechanism, the scientific literature does offer a precise definition of ‘pathway’, and hence we might have a sounder grasp of its epistemological import in scientific practice.
One of the best known and most widely used databases, i.e. Gene Ontology, “the framework for the model of biology”, “defines concepts/classes used to describe gene function, and relationships between these concepts. It classifies functions along three aspects: molecular function: molecular activities of gene products; cellular component: where gene products are active; biological process: pathways and larger processes made up of the activities of multiple gene products” (http://www.geneontology.org/). Although the Dictionary of Cancer Terms of the National Cancer Institute (NCI) does not have any definition of molecular mechanism, we find a precise and unambiguous definition of molecular pathway: “A series of actions among molecules in a cell that leads to a certain end point or cell function” (https://www.cancer.gov/publications/dictionaries/cancer-terms?cdrid=561605). It is noteworthy that this definition stresses four different and highly relevant aspects: (1) any pathway is the representation of a series of causal actions among molecules; (2) any pathway is the representation of a series of actions among molecules that end in a particular function (and molecular biologists—as we know very well—are extremely interested in the function of molecules, in particular in a DNA or RNA sequence or in a protein); (3) any pathway is the representation of a number of actions among molecules, where such a number can vary from n = 1 (there is just one action leading to one molecular bond) to n = m (there are m bonding steps, depending on the length of the pathway considered); (4) any pathway is the representation of a temporally continuous process of bonding steps.
To further improve our understanding of how scientists really work in this area, we should be aware that while we do not have any widely shared database on molecular mechanisms, we have many extremely widely used databases on pathways, like, for example: the Kegg Pathway (http://www.genome.jp/kegg/pathway.html); the Small Molecule Pathway Database (http://smpdb.ca/); the Reactome (https://reactome.org/); the ConsensusPathDB-human (http://cpdb.molgen.mpg.de/); the Pathguide (http://www.pathguide.org/); WikiPathways (https://www.wikipathways.org/index.php/WikiPathways); and others. It seems therefore that, if philosophers of science aim to account for the epistemological features of scientific practice, pathways must be taken into account at least as much as mechanisms—and maybe more.
Shifting our attention from just mechanisms to pathways also means addressing another aspect of the molecular biology literature: both the titles and texts of many papers refer to both mechanisms and pathways, thus proving that authors consider them two different things.Footnote 7 If this is the case, then we seem to be entitled to conclude, at variance from Ioannides and Psillos, that mechanisms and pathways do not coincide, with mechanisms being used by scientists in a fairly vague and loose way, and pathways in a precise and clear way, as series of continuous steps in molecular processes.
A focus on pathways brings with it a shift in conceiving explanation as well: it implies a focus on the performance of a specific function, which is enacted by virtue of specific molecular set-up conditions, i.e., of a specific molecular context. For each pathway, “we can specify the set-up conditions as the molecules that initiate the first reaction in the pathway, and the terminating conditions as the molecules that are produced by the last reaction in the pathway” (Thagard 2002, p. 238). Changes in the molecular context constrain the performance of the function which that specific pathway is meant to act out and, of course, our epistemological understanding of such a function. Different contexts allow for different functional explanations of the behaviour of the same molecule. In other terms, the specific context of instantiation of the pathway’s acting conditions the epistemic validity of its explanation in functional terms. While not incompatible with mechanism, this perspective suggests a different weighting of the notion of functionFootnote 8 and of the context in which it is performed—where the context could be thought of as defining the conditions of adequacy of the explanation itself.
Recalling some examples from molecular biology
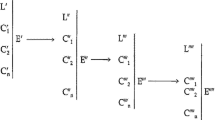
In order to move from philosophical reflections to real science, let us consider some cases that could support the claims above, and spur a deeper insight into pathways. To better address this task, we introduce a few examples and a specific notation to convey them that can be useful for our aims. Actually this notation has been proposed for text mining, automatic discovery and for connecting system biology and molecular biology.Footnote 9 To apply it effectively here, we leave aside some of its more technical aspects, and use the most intuitive and useful for our aims.
Let us consider one of the several pathways that compose the metabolic network, in particular the pathway that starts with d-glucose (Glc) and ends with fructose-1,6-bisphosphate (F1, 6P). As in nearly all cases, this pathway contains molecules that are collected from the cellular or extracellular environment, and molecules that are produced in the course of the pathway itself. In our case, the molecules from the environment are Glc, hexokinase (HK), glucose-6-phosphate isomerase (GPI), 6-phosphofructokinase (PFK), and two adenosine triphosphates (ATP). The intermediate product is d-glucose-6-phosphate (G6P), while the final one is fructose-6-phosphate (F6PP). Let us group together the molecules from the environment, and use & to indicate their collection, that is, Glc&HK&GPI&PFK&ATP&ATP. Therefore, we can indicate the pathways by Glc&HK&GPI&PFK&ATP&ATP ├ F1,6P, and the component causal steps by
1. Glc & HK → Glc*HK |
2. (Glc*HK) & ATP → (Glc*HK)*ATP |
3. (Glc*HK) *ATP → G6P & HK & ADP |
4. G6P & GPI → G6P*GPI |
5. G6P*GPI → F6P & GPI |
6. F6P & PFK → F6P*PFK |
7. (F6P*PFK) & ATP → (F6P*PFK) *ATP |
8. (F6P*PFK) *ATP → F1,6P & PFK & ADP |
where * indicates a bond between what is at its left side and at its right side; instead → indicates the singular causal step from certain molecules (on the left) to their product (on the right).
From this example we grasp some important aspects, already highlighted by the NCI definition of pathway. First of all, each pathway “lives” in a molecular context which is defined by a set of molecules (in our case, Glc&HK&GPI&PFK&ATP&ATP) and which identifies the function of any molecule involved. We will come back to this issue below. Moreover, there is no pathway if any step is not due to a causal interaction (in turn due to thermal agitation) among the molecules involved. Such an interaction, caused by thermal agitation, ends in a bond which is also facilitated by the three-dimensional configuration of the binding molecules and, thus, by their stereospecificity. Moreover, the sequence of the steps (eight in the case above) is continuous in time.
A further important issue is that we could consider only the shortest pathway (i.e., Glc&HK ├ Glc*HK), which is represented by a two-molecule context (Glc&HK) and by one product given by their compound (Glc*HK). Now we clearly have only one causal step:
1. Glc & HK → Glc*HK |
But we can also ask for the functional explanation of the molecule Glc. In this case, and given that molecular context, the answer is very simple, that is, the functional explanation of Glc is given in terms of its bond with HK. We could name this immediate function of the molecule its direct function. Of course, any molecule has a direct function, which is given by the minimal pathway in which it is located. It should be noted that if we have the same molecule belonging to two different minimal pathways (that is, if it has the possibility to bind to two different molecules), then it has two different direct functions, as we will exemplify later on.
Let us go on and consider the Glc&HK&ATP ├ (Glc*HK) *ATP pathway, which “lives” in a molecular context given by Glc&HK&ATP. This time there are two causal steps in the pathway:
1. Glc & HK → Glc*HK |
2. (Glc*HK) & ATP → (Glc*HK) *ATP |
If now we ask for the functional explanation of Glc, we have to pay some attention, since it is twofold: there is a direct function (the bond with HK), but there is also a final function (the production of the compound Glc*HK*ATP).
We can proceed and consider the Glc&HK&ATP ├ G6P pathway, which “lives” in the molecular context Glc&HK&ATP and which is composed of three causal steps:
1. Glc & HK → Glc*HK |
2. (Glc*HK) & ATP → (Glc*HK) *ATP |
3. (Glc*HK) *ATP → G6P & HK & ADP |
Again, the direct function of Glc is the bond with HK, but now the final function of Glc is the production of G6P.
Thus, to generalise, let us stress that in any pathway each molecule involved has a direct function, which is given by its bond with the molecule/s with which it produces a compound, and a final function, which is understood considering the pathway in its entirety. We have already pointed to the relativity of the notion of direct function, since the same molecule can bind to different compounds. But even the notion of final function can be deemed relative, since it depends on the pathway taken into consideration. Of course, if the pathway is composed of only one step, the direct function and the final function are the same. A molecule can play many functions, depending on the pathway in which it is involved. We can also introduce the notion of the main function, which could be thought of as the final function of a molecule with respect to the pathway, the net of pathways or even the entire organism that scientists consider the relevant starting point (some examples are provided below).
Considering the succession of pathways in which the given molecule is inserted (Glc&HK ├ Glc*HK; Glc&HK&ATP ├ (Glc*HK) *ATP; Glc&HK&ATP ├ G6P; etc.), we can grasp the different functions it could have. But there is an important point to consider here: in order to understand the main function of that molecule, we have to find the right pathway. For example, if we want to investigate (and thus explain) the main function of the Glc molecule, we cannot stop at the third pathway above, but have to proceed up to what is called the metabolic pathway (and that is actually a net of pathways), which includes the eight steps above but continues and ends with the production of pyruvate and the release of ATP. It is from this longer pathway—which gives us the process of glycolysis—that we understand that the main function of Glc is to serve as a primary source of energy for all living organisms. In this case the attribution of the title of main function to Glc depends on the fact that we have considered the organism level.
There is, however, another point worth stressing. Even if the pathway can be thought of as composed of n causal steps, it cannot be epistemically decomposed into those n steps. The reason rests on the fact that the function of the first molecule of the pathway, that is, of the molecule from which the pathway starts, is strictly linked to the final molecule of the pathway, that is, it depends on the entire pathway. In other terms, the function of a molecule cannot be decomposed. If we take a shorter or longer pathway, as illustrated above, we have to attribute it a different function; but it is the entire pathway under consideration that provides us with the epistemological understanding of its function. In particular, if we want to grasp the main function, for example, of Glc, we have to consider the entire metabolic pathway (and then the entire organism), and cannot epistemically decompose it: the molecule’s function of being a source of energy can be explained only if we examine the entire pathway, and not if we consider component subpathways.
In order to grasp another epistemic feature of the molecular context, let us go back to two subpathways of the metabolic pathway already considered, that is, Glc&HK&ATP ├ (Glc*HK) *ATP and Glc&HK&ATP ├ G6P. As we can see, the respective molecular contexts are the same. This means that there could be situations in which the identification of the molecular context does not disambiguate the pathway and therefore the functional role of the molecule under investigation. Therefore, while the molecular context identifies the set of possible functional explanations, it is the pathway that fixes the right one.
Let us move to a different instructive example. We know that TP53 is a protein, encoded by the TP53 gene located on the short arm of human chromosome 17, whose main function is being a tumour suppressor. If we want to investigate this function, we have to identify the entire tumour suppressor pathway and its molecular context. This protein, indeed, is a central node in a complex signalling pathway, evolved to detect a variety of cytotoxic and genotoxic stresses that could compromise genomic stability and promote neoplastic transformation. Once activated by a stress signal such as DNA damage, hypoxia, unscheduled oncogene expression, viral infection, or ribonucleotide depletion, TP53 exerts its function of “guardian of the genome” and mediates a series of cellular outcomes that can vary from cell cycle arrest to DNA-repair, senescence and apoptosis, depending on the cellular context. This means that if TP53 is mutated its main function of tumour suppressor no longer works, and this can end in several kinds of cancers.
If we consider TP53 inside the mentioned signalling pathway, we realise that it has the function of being a tumour suppressor. But TP53 is also inside a different pathway, and if we consider this we have to reach the conclusion that it also has a different function. Let us grasp this point better. TP53 is also a central molecule of a regulation pathway ending with its own degradation. Here there is the ubiquitin ligase MDM2, a protein encoded by the gene MDM2, which binds with TP53, thus inhibiting its transcriptional activity and stimulating its ubiquitination, that is, its bond with ubiquitin (U), and consequent proteasome-dependent degradation (call it degTP53). In other words, ubiquitination is the process that causes a protein, in this case TP53, to be labelled by the protein ubiquitin. This labelling step marks TP53 for destruction by proteasomes (P), very large protein complexes within the cell. Summing up, TP53 also has the function of regulating the activation of a protein (MDM2), which is itself a regulator of TP53. This regulation pathway can be described as follows:
TP53& TP53& MDM2& U&P ├ degTP53
And the causal steps are:
1. TP53 & MDM2 → TP53*MDM2 |
2. TP53 * MDM2 → MDM2 |
3. MDM2 & TP53 → MDM2*TP53 |
4. (MDM2 *TP53) & U → (MDM2*TP53) *U |
5. (MDM2*TP53) *U → MDM2 & (TP53*U) |
6. (TP53*U) & P → (TP53*U) *P |
7. (TP53*U) *P → degTP53 & U & P |
Thus, if we want a functional explanation of TP53, we have to identify the particular pathway in which it is inserted. If we want to understand its function as a tumour suppressor, we have to consider the right signalling pathway; if we want to understand its function as a self-regulator, we have to consider the TP53&TP53&MDM2&U&P ├ dTP53 pathway and thus the related molecular context: TP53&TP53&MDM2&U&P.
From this example, it is again clear that, even if the pathway can be thought of as composed of causal steps (in this case, seven), it cannot be epistemically decomposed since the regulatory function of the molecule (TP53) or its being a tumour suppressor can be explained only by the related pathways in their entirety. This is why only the notion of pathway as a continuous causal process can capture crucial epistemological features both of the molecule under analysis and of the molecular constraints under which it operates.
Further aspects must be emphasized. By examining the molecular context of the pathway under consideration, we are also able to explain the function of other molecules involved: the function of MDM2 is to inhibit the transcriptional activity of TP53 and to facilitate the ubiquitination (the bond with ubiquitin); the function of ubiquitin is to label the molecule (in this case, TP53) that has to be degraded; the function of the proteasome is to degrade the ubiquinated protein (in this case, TP53). But, again, we are able to attribute these functions only by examining the pathway in its entirety.
Let us add another example showing that the same molecule—SCF: a growth factor (GF in the figure below)Footnote 10—has two different final functions, since it belongs to two different pathways (the MAPK and PI3 K signalling pathways belonging to the KEGG melanoma pathway; see figure below taken from www.genome.jp/kegg/pathway/hsa/hsa05218.html, and Boniolo and Lanfrancone 2016).Footnote 11 Actually in this case, the starting molecules are the same three: SCF, c-KIT (RTK in the figure) and RAS (Rat sarcoma protein).Footnote 12

The MAPK signalling pathway is central in regulating many cellular processes, including inflammation, cell stress response, cell differentiation, cell division, cell proliferation, metabolism, motility and apoptosis. It is composed of a chain of causal steps communicating a signal from a receptor on the surface of the cell to the DNA in its nucleus. The pathway has a major role in many pathologies, since, when one of the proteins involved is mutated, it can become stuck in the “on” or “off” position, which is a necessary step in the development of cancers, immune disorders and neurodegenerative diseases.
Instead, the PI3K-Akt signalling pathway is involved in regulation of the cell cycle. It plays a central role, especially with respect to many cancers (melanoma in particular), since cell cycle regulation has to do with cell growth, survival, motility, and metabolism. As noted, they both start from SCF, and the bond between SCF and c-KIT leads to c-KIT dimerization and autophosphorylation, thereby leading to the activation of RAS. Independently of the biological meaning of the steps and the presence of intermediate molecules, at the end we have two pathways: according to one (the MAPK pathway) SCF’s final function is the increase in melanocytic cells; according to the other (the PI3K-Akt pathway) SCF’s final function is the regulation of MITF.
To conclude this illustrative part, we wish to recall the important case of drugs and how the focus on certain pathways also plays a central role in this setting. As is known, drugs are molecules that change the molecular context of certain “non normal” or “pathological” pathways, either inhibiting certain “non normal” or “pathological” functions or activating new anti-symptomatic or curative functions. We cannot explain these functions without taking into consideration the pathways in which they are inserted and how, after this insertion, the molecular context changes and, therefore, so does the function of the molecules involved. Let us consider thermoregulation in humans. We know that it is a feedback control pathway maintaining body temperature at around 37 °C. For many reasons, the temperature can increase perilously. In this case we take paracetamol (a drug) to control it. This is an inhibitor. As such, it inhibits the cyclooxygenase pathway, which is responsible for formation of prostanoids. In particular, like any drug, paracetamol can be considered an ad hoc external molecule inserted in an already existing internal pathway (in this case, the thermoregulation feedback pathway) in order to interrupt the “non-normal” or “pathological” process of a cell (or of a cell population) in some way, by attempting to divert it from its “natural history”. Most antibiotics work in this way. An example is given by those whose active element (as in rifampicin or rifampin) is a protein synthesis inhibitor. This either stops or slows down the growth or the proliferation of the infecting bacteria by disrupting the pathways involved in the production of their proteins. Passing to HIV infection, the disease (but also some cancers) can be treated by an antiretroviral drug based on the reverse-transcriptase inhibitors. The drug inhibits the pathways involved in the activity of a viral DNA polymerase enzyme (that is, the reverse transcriptase) used by the retroviruses to reproduce. After the virus has infected a cell, the reverse transcriptase copies the single-stranded viral RNA into a double-stranded viral DNA. This is then integrated into the host DNA, thereby allowing host cellular processes to reproduce the virus. The reverse-transcriptase inhibitors stop the reverse transcriptase activity and prevent the synthesis of the double-stranded viral DNA, thus preventing HIV from multiplying. It is worth noting that sildenafil, used to cure male erectile dysfunction, also works in this way by inhibiting cGMP-specific phosphodiesterase type 5, i.e. the enzyme involved in the degradation pathway of cyclic guanosine monophosphate. This is a signalling molecule triggering smooth muscle relaxation and allowing blood flow into the corpus cavernosum, which causes erection. As an effect of the decrease of the cGMP-specific phosphodiesterase activity due to sildenafil, the degradation of the signalling molecule is stopped and, consequently, this signal lasts for a longer period of time.
This wide array of examples can be taken as representative of the use of causal and functional notions in molecular biology, and provides us with hints regarding explanatory strategies in that field, in particular concerning how central the notion of pathway is to understanding what really happens both from the scientific and the epistemological points of view. Our philosophical reflections, which will be further developed in the following section, are thus rooted in scientific literature, and find their direct counterpart in an array of recent works in molecular biology addressing context-dependence (see e.g. Schwanbeck et al. 2011; Egloff and Grandis 2012; Lan et al. 2013; Bray 2016).
The explanatory centrality of molecular pathways
What has been said up to now should strongly indicate that the notions of process and of the context in which it “lives” have to be retrieved and deserve more attention in a philosophical discourse on the biological scenario, especially if we are focusing on the molecular level. Neo-mechanistic theories have turned away from processual accounts but this move has probably not been totally fruitful. Some twist in the current philosophy of science debate can benefit from paying attention to processes, in the very spirit of drawing closer to scientific practice that largely motivates it.
Dealing with cell biology and, more specifically, with mitosis, Dupré (2013) remarks for instance how “the formation of the microtubules that constitute the mitotic spindle, which in turn separates the two strands of DNA in the chromosomes of the dividing cell, […] is, surely, a biological function. Yet it turns out that it does not individuate a mechanism. The reason for this is just that there are a number of different ways in which microtubule growth (or degradation) is directed; if one of these is prevented by some kind of intervention, then other mechanisms take up the slack” (p. 26).Footnote 13 Along similar lines, the examples provided above take these claims a few steps further, referring to different ways in which molecular functions can be enacted and to the crucial role and impact of the molecular context.
Our analysis, which is confined to molecular biology, has aimed to show that neo-mechanism, while already constituting a very broad and developed philosophical movement, still fosters further reflections and clarifications of concepts used in the sciences. The latter, though, might not be most effectively concerned with some refinement of the notion of mechanism in terms of entities and activities—as largely pursued in the latest literature—but, rather, on some rethinking of ‘process’ and ‘pathway’, as suggested by accounts which preceded neo-mechanism and which we believe rightly stressed continuity of production as a core feature of causal relations.
At this point, a remarkable asymmetrical situation is worth recalling: on the one hand, the biomedical sciences have no definition of mechanism, while philosophy of science has many of definitions of mechanism; on the other, the biomedical sciences have clear and basically convergent definitions of pathway, while philosophy of science has paid much less attention to this element, which lies at the core of molecular biology as it allows us to grasp the functions of the molecules we are interested in. To help overcome this deficiency, and starting from the NCI’s definition, we could state that
A molecular pathway is continuous series of n causal bonding steps starting from one molecule and leading to a molecular outcome with a certain function, which is determined by the entire pathway itself. Any molecular pathway “lives” in a given molecular context composed of a certain set of molecules that fixes the conditions of possibility of its enactment.
The examples illustrate the crucial explanatory role of the molecular context which sets the conditions of enactment of the molecular pathway and plays a decisive role in the explanation of molecular functioning.
We should now go back to where we started from, that is, the success of the new mechanical philosophy and its ambition to make sense of causal discourses in the sciences. In the light of the examples, we should wonder what role mechanisms play in molecular biology, and question whether they are necessary and/or sufficient.
As it appears, the notion of mechanism is not necessary to describe the molecular pathways referred to above: the illustration of their functioning has made no epistemological appeal to mechanisms. On the one hand, while not incompatible with mechanistic accounts, the analysis provided does not depend on any specific proposal on how a mechanism is to be conceived, or how a mechanistic model is to be built. On the other hand, current definitions of mechanism like the ones mentioned in “Mechanisms and the new mechanical philosophy: What’s new?” section—i.e. Illari and Williamson’s and Glennan’s—seem too general and broad to effectively capture what goes on in the molecular actions at stake. The notions of pathway, function and causal interaction do the job, by expressing, if properly understood, both the causal aspect inherent in molecular processes—namely, the production of a given outcome—and the functional one, together with a proper stress on the continuity of the molecular actions and their contextual constraints.
What about other philosophical definitions of mechanism? Do they do any better in the respects at stake here? Let us consider two of the most successful ones, which have already turned more than one decade, are still widely discussed, and do actually take functions into serious account. According to Machamer, Darden and Craver, “mechanisms are identified and individuated by the activities and entities that constitute them, by their start and finish conditions, and by their functional role. Functions are the roles played by the entities and activities in a mechanism. To see an activity as a function is to set it as a component in some mechanism, that is, to see it in a context that is taken to be important, vital or otherwise significant. […] Functions should be understood in terms of the activities by virtue of which entities contribute to the workings of a mechanism” (Machamer, Darden and Craver 2000, p. 6). While far from being opposed to this definition, our view is significantly different in a few respects. To start with, we believe that an entity ontology—whichever entities we bottom-out on—is not the most suitable starting point in molecular biology in cases like the ones we analysed, which are not best thought of in terms of discrete elements or decomposability of systems. Once we replace entities with pathways, continuity—an aspect neo-mechanists are not overconcerned with—can gain due appreciation. Furthermore, the appeal to pathways seems to do justice to the distinctive contribution of the molecular arrangement itself (what we deemed the “molecular context”), which is thoroughly acknowledged.
Things seem somewhat trickier if we look at Bechtel and Abrahamsen’s definition: “A mechanism is a structure performing a function in virtue of its component parts, component operations, and their organization. The orchestrated functioning of the mechanism is responsible for one or more phenomena” (2005, p. 423). Although organized behaviour and the performing of functions are properly recognised, here too the focus remains on components, “because [the authors] want to draw attention to the involvement of parts”, with actually some concern for accounts focusing on linear processes (ibidem, footnote 5). In the cases we have analysed, the real interest is not merely in which units are at stake, but in the continuous processual character of the pathways: the only ground on which we can propose a functional explanation of a molecule. To better illustrate this point, in addition to the examples already given, let us think about the protein cytochrome complex (Cyt c). Usually located in the mitochondrial intermembrane space, Cyt c is also found in the cytosol. In the intermembrane space Cyt c is inserted in a particular pathway; in the cytosol in a totally different one. Focusing on the intermembrane space, we explain Cyt c’s function in terms of cellular respiration; focusing on the cytosol, we explain its function in terms of cellular apoptosis! Summing up, we need pathways to explain the possible multifunctional roles of a molecule. Mechanisms seem not to be enough. Moreover, mechanisms do not seem to be adequate in helping us to understand that a molecule can have as many functions as the many subpathways starting from that molecule. But we have also seen that there is only one pathway that allows us—as molecular biologists—to attribute to that molecule what we think is its main function. Recalling the example above, the main function of Glc d-glucose is not to bind with HK, or produce G6P, or F6PP, etc., but to serve as a source of energy. And we understand this only when we consider the pathway starting from Glc d-glucose and ending with pyruvate.
Furthermore, issues regarding gaps in our knowledge of mechanistic systems are dealt with differently here. If Salmon, as well as, e.g., Peter Railton, already acknowledged that in the great majority of cases we do not get to know mechanisms exhaustively, completely and in full detail, the issue has been more thoroughly addressed by neo-mechanists. Machamer et al. (2000), e.g., have stressed how we mostly arrive at mechanisms’ sketches or schemata, and Craver (2006) has distinguished between how-possibly, how-plausibly and how-actually mechanistic models. At the same time, in order to grasp the heuristics guiding the elaboration of mechanistic models in the life sciences, Bechtel and Richardson (1993) have emphasized the virtues of structural and functional decomposition. We take the examples provided in the paper to show that the notion of pathway is central to navigate cases in which the purpose is not to express some gap to be epistemically filled, or to present some systems to be decomposed. The core elements that scientific explanations in molecular biology are called to unravel in cases like those illustrated above are continuous pathways, and their functions, which we currently know a good deal about. What we aim to stress is that the definition of pathway provided above improves our attempts to grasp the performing of a certain function, the way in which this is done (as a continuous series of causal bonds), and its internal constraints.
From processes to mechanisms, and back
Let us conclude by highlighting how a focus on the notion of pathway with respect to the analysis of causation in molecular biology can bring a few take-home lessons with it, both with respect to the specific field at stake, and with respect to reflections on mechanisms and related concepts. In the first place, the notion of pathway ties the knot between talking of mechanisms and talking of functions. Neo-mechanists have pointed out, for instance, that “mechanisms are functionally individuated by their phenomena” (Illari and Williamson 2012, pp. 123–124), or that functional explanations—specifically, in psychology—can only be conceived as sketchy mechanistic accounts (Piccinini and Craver 2011). We stress how the context proves crucial to the very enacting of the molecular pathway itself, and hence to the proper performing of its function and the bringing about of a given outcome. Outcomes of molecular processes, in other terms, are brought about by pathways whose correct functioning is strictly conditional upon particular molecular features. It is in this sense that we suggest an adequate explanation of the working of molecular pathways must be contextual.
Therefore, the epistemological considerations we have elaborated starting from the scientific framework recalled above also involve some rethinking of the notion of context and its role in tackling mechanisms. Already in the late Salmon, the context is taken into account as impacting on the identification of causal nexus. While remaining faithful to his ontic conception, Salmon admits that “statements about the relations between causes and effects are usually highly selective, and they are typically context-dependent” (Salmon 2002, p. 126). Basically following like intuitions, in the scenario of the new mechanical philosophy the context has been held to guide the process of mechanisms’ discovery,Footnote 14 with background knowledge and interests playing a part: in biology, “first, the choice of phenomenon is relative to the scientist’s interests. […]. To some extent and in some cases, the choice of beginning, ending, topping-off, and bottoming-out points in the description of a mechanism may also be related to the interests of the investigator” (Darden 2008, p. 960). While agreeing with these claims, we believe that the context is to be seen here as not hinting only at external contingent conditions, and thus not only performing a role with respect to our epistemic practices, in the construction of our mechanistic models. The context can be taken as decisive to the very functioning of some processes the way they do, as related to the very configuration of the phenomenon under investigation: there is no biological process if not in a given biological context. This means that if the biological context changes, the biological process changes as well. And a pathway approach, like the one we are proposing here, puts this point right at the centre. We hence suggest that an additional notion of context is to be introduced, which has not to do with the way in which the elaboration of our causal knowledge is constrained by pragmatic factors, but with the very conditions allowing biological functions to be enacted. When discussing explanatory matters, it is to be conceived both as a “perspectival affair” (Craver 2001, p. 73), related to our epistemic procedures, and, equally (or more) importantly, as an aspect inherent to the conditions of acting of the pathway itself, which an explanation of molecular behaviour must account for. That is what, we believe, the notion of pathway can capture better than currently available notions of mechanism.
Without denying the many merits of neo-mechanism, we claim that, when wondering which notions provide the most valuable guidance in tackling productive and explanatory relations in the sciences, we should not discount or bracket the notions of process and pathway too quickly. Dealing with very specific and detailed examples in molecular biology can help us re-assess the epistemological import of both the notions of mechanism put forward by neo-mechanists, and the notions of process/pathway and contexts in which they “live”. All in all, if the purpose of philosophical enquiry on causal discourse in the sciences is to provide the fittest conceptual tools to facilitate and support scientific investigation—a daunting challenge indeed—we should wonder whether the struggle for a one-fits-all notion of mechanism is the most promising route to follow, or conversely, whether we should pay more attention to the processes and the contexts allowing their enactment.
Notes
A wide survey of neo-mechanism as a philosophical movement is provided in Glennan and Illari (2017a).
Such features as isolability, organization and sequentiality are taken to situate processes along some “multidimensional gradient”. This is opposed to an analysis on the basis of discrete entities (Skillings 2015, p. 1140).
Discussing biomedical discovery, Paul Thagard too states: “biochemical pathways are a kind of mechanism. […] In biochemical pathways, the entities are the molecules and the activities are the chemical reactions that transform a molecule into other molecules” (2002, pp. 237–238, italics added).
According to Ioannides and Psillos, the three major metaphysical accounts of mechanisms nowadays are Machamer et al. (2000), Glennan’s (2002) and Bechtel and Abrahamsen’s (2005). We already introduced Glennan’s most recent definition above, and will refer to the other two works below. Psillos’ work on mechanistic causation has not only addressed most recent theories and trends, but also thoroughly investigated the historical roots of the philosophy of mechanisms. See e.g. Psillos (2011).
Dowe’s account has been amended in Boniolo et al. (2011).
On the suggestion of an overall rethinking of the living world in processual terms, up to a renewed philosophy of biology, see Dupré and Nicholson (forthcoming). For some arguments against the metaphysical replacement of mechanisms with processes, see Austin (2016).
We are far from denying that functions have also been addressed within philosophical discussions of mechanisms. Just to recall some very recent work, Justin Garson (2017), e.g., stresses that mechanisms are sometimes described in terms of their functions. However, he remains faithful to the notion of mechanism: “the class of functional mechanisms is [taken as] an interesting proper subset of the class of minimal mechanisms” (Garson 2017, p. 105), where “minimal mechanisms” are understood according to Glennan’s (2017) definition. We hence believe that this analysis is not immune to the shortcomings of the far too broad notion of minimal mechanism.
As is known, many genes and the encoded proteins have different names. We follow the nomenclature indicated by the HUGO Gene Nomenclature Committee, (www.genenames.org).
Melanoma, as is known, is a type of skin cancer. MAPK is a mitogen-activated protein kinase (originally called ERK—extracellular signal-regulated kinase—as in the figure) and PI3 K is another protein: phosphatidylinositol-4,5-bisphosphate 3-kinase.
c-KIT is a tyrosine kinase receptor and RAS (Rat sarcoma protein) is a serine/threonine-protein kinase.
Commenting on that, Dupré continues: “The problem with this kind of teleological system, or system with a robust tendency to end up in particular preferred states, is that the relation between functions and mechanisms is more complex than that supposed by standard mechanistic theories” (ibidem).
Amongst most recent works on this, see e.g. Darden et al. (2018).
References
Austin CJ (2016) The ontology of organisms: Mechanistic models or patterned processes? Biol Philos 31:639–662
Bechtel W, Abrahamsen A (2005) Explanation: mechanist alternative. Stud Hist Philos Biol Biomed Sci 36:421–441
Bechtel W, Richardson R (1993) Discovering complexity. Princeton University Press, Princeton
Boniolo G, Lanfrancone L (2016) Decomposing biological complexity into a conjunction of theorems. The case of the melanoma network. Humana.Mente. J Philos Stud 30:19–35
Boniolo G, Di Fiore PP, D’Agostino M (2010) Zsyntax: a formal language for molecular biology with projected applications in text mining and biological prediction. PLoS ONE 5:e9511. https://doi.org/10.1371/journal.pone.0009511
Boniolo G, Faraldo R, Saggion A (2011) Explicating the notion of ‘causation’: the role of the extensive quantities. In: Illari P, Russo F, Williamson J (eds) Causality in the sciences. Oxford University Press, Oxford, pp 502–525
Boniolo G et al (2013) A logic of non-monotonic interactions. J Appl Logic 11:52–62
Boniolo G et al (2015) Adding logic to the toolbox of molecular biology. Eur J Philos Sci 5:399–417
Bray SJ (2016) Notch signalling in context. Nat Rev Mol Cell Biol 17:722–735
Craver C (2001) Role functions, mechanisms and hierarchy. Philos Sci 68:53–74
Craver C (2006) When mechanist models explain. Synthese 153:355–376
Craver C, Darden L (2013) In search of mechanisms. University of Chicago Press, Chicago
Darden L (2008) Thinking again about mechanisms. Philos Sci 69:S354–S365
Darden L et al (2018) The product guides the process: discovering disease mechanisms. In: Danks D, Ippoliti E (eds) Building theories. Springer, Dordrecht, pp 101–118
Dowe P (2000) Physical causation. Oxford University Press, Oxford
Dupré J (2013) Living causes. Proceedings of the Aristotelian Society Supplementary Volume LXXXVII, https://doi.org/10.1111/j.1467-8349.2013.00218.x
Dupré J, Nicholson D (forthcoming) A manifesto for a processual philosophy of biology. In: Nicholson D, Dupré J (eds) Everything flows: towards a processual philosophy of biology. Oxford University Press, Oxford
Egloff AM, Grandis JR (2012) Molecular pathways: context-dependent approaches to Notch targeting as cancer therapy. Clin Cancer Res 18(19):5188–5195. https://doi.org/10.1158/1078-0432.ccr-11-2258
Franklin-Hall L (2016) New mechanistic explanation and the need for explanatory constraints. In: Aizawa K, Gillett C (eds) Scientific composition and metaphysical ground. Palgrave MacMillan, Basingstoke, pp 41–74
Garson J (2017) Mechanisms, phenomena, and functions. In: Glennan S, Illari P (eds) The Routledge handbook of mechanisms and mechanical philosophy, pp 104–103
Giovannetti E et al (2013) Molecular mechanisms and modulation of key pathways underlying the synergistic interaction of sorafenib with erlotinib in non-small-cell-lung cancer (NSCLC) cells. Curr Pharm Des 19:927–939
Glennan S (2002) Rethinking mechanistic explanation. Philos Sci 69:S342–S353
Glennan S (2017) The new mechanical philosophy. Oxford University Press, Oxford
Glennan S, Illari P (eds) (2017a) The Routledge handbook of mechanisms and mechanical philosophy. Routledge, London
Glennan S, Illari P (2017b) Varieties of mechanisms. In: Glennan S, Illari P (eds) The Routledge handbook of mechanisms and mechanical philosophy. Routledge, London, pp 91–103
Hitchcock C (2004) Causal processes and interactions: What are they and what are they good for? Philos Sci 71:932–941
Illari P, Williamson J (2012) What is a mechanism? Thinking about mechanisms across the sciences. Eur J Philos Sci 2:119–135
Ioannidis S, Psillos S (2017) In defense of methodological mechanism. Axiomates 27:601–619
Kimball SR, Jefferson LS (2016) Signaling pathways and molecular mechanisms through which branched-chain amino acids mediate translational control of protein synthesis. J Nutr 136:227S–231S
Lan A, Ziv-Ukelson M, Yeger-Lotem E (2013) A context-sensitive framework for the analysis of human signalling pathways in molecular interaction networks. Bioinformatics 29:i210–i216
Levy A (2013) Three kinds of new mechanism. Biol Philos 28:99–114
Machamer P, Darden L, Craver C (2000) Thinking about mechanisms. Philos Sci 67:1–25
Nethi SK et al (2015) Investigation of molecular mechanisms and regulatory pathways of pro-angiogenic nanorods. Nanoscale 7:9760–9770
Nicholson D, Dupré J (eds) (forthcoming) Everything flows: towards a processual philosophy of biology. Oxford University Press, Oxford
Nosrati N et al (2017) Molecular mechanisms and pathways as targets for cancer prevention and progression with dietary compounds. Int J Mol Sci 18:2050
Piccinini G, Craver C (2011) Integrating psychology and neuroscience: functional analyses as mechanism sketches. Synthese 18:283–311
Psillos S (2011) The idea of mechanism. In: Illari P, Russo F, Williamson J (eds) Causality in the sciences. Oxford University Press, Oxford, pp 771–788
Rosenberg A (2018) Making mechanism interesting. Synthese 195:11–33
Salmon W (1984) Scientific explanation and the causal structure of the world. Princeton University Press, Princeton
Salmon W (1989) Four decades of scientific explanation. In: Kitcher P, Salmon W (eds) Scientific explanation, vol XIII. Minnesota Studies in the Philosophy of Science, Minneapolis, pp 3–219
Salmon W (1998) Causality and explanation. Oxford University Press, Oxford
Salmon W (2002) A realistic account of causation. In: Marsonet M (ed) The problem of realism. Ashgate, Aldershot, pp 106–134
Schwanbeck R, Martini S, Bernoth K (2011) The Notch signaling pathway: molecular basis of cell context dependency. Eur J Cell Biol 90:572–581
Skillings DJ (2015) Mechanistic explanation of biological processes. Philos Sci 82:1139–1151
Thagard P (2002) Pathways to biomedical discovery. Philos Sci 70:235–254
Wang Z et al (2016) The molecular mechanism and regulatory pathways of cancer stem cells. Cancer Transl Med 2:147–153
Acknowledgments
We would like to thank the referees and J. Dupré for their very useful comments on a previous version of this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Boniolo, G., Campaner, R. Molecular pathways and the contextual explanation of molecular functions. Biol Philos 33, 24 (2018). https://doi.org/10.1007/s10539-018-9634-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10539-018-9634-2