Abstract
Selenium (Se) plays an essential role in the growth of fish and performs its physiological functions mainly through incorporation into selenoproteins. Our previous studies suggested that the selenoprotein W gene (selenow) is sensitive to changes in dietary Se in rainbow trout. However, the molecular characterization and tissue expression pattern of selenow are still unknown. Here, we revealed the molecular characterization, the tissue expression pattern of rainbow trout selenow and analyzed its response to dietary Se. The open reading frame (ORF) of the selenow gene was composed of 393 base pairs (bp) and encodes a 130-amino-acid protein. The 3′ untranslated region (UTR) was 372 bp with a selenocysteine insertion sequence (SECIS) element. Remarkably, the rainbow trout selenow gene sequence was longer than those reported for mammals and most other fish. A β1-α1-β2-β3-β4-α2 pattern made up the secondary structure of SELENOW. Furthermore, multiple sequence alignment revealed that rainbow trout SELENOW showed a high level of identity with SELENOW from Salmo salar. In addition, the selenow gene was ubiquitously distributed in 13 tissues with various abundances and was predominantly expressed in muscle and brain. Interestingly, dietary Se significantly increased selenow mRNA expression in muscle. Our results highlight the vital role of selenow in rainbow trout muscle response to dietary Se levels and provide a theoretical basis for studies of selenow.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Selenium (Se) is an essential micronutrient, which is involved in a number of pathological and physiological functions in vertebrates. Se deficiency can cause serious diseases in animals and humans, such as white muscle disease (WMD), keshan disease and cardiovascular disease (Rayman 2000). Moreover, Se mainly functions in the form of selenocysteine (Sec) through selenoproteins (Gladyshev 2001). Sec is known as the 21st amino acid (aa) in protein, which has its own codon (UGA) and Sec-specific biosynthetic as well as insertion machinery (Hoffmann and Berry 2005). Currently, more than 50 selenoproteins have been identified across the three domains of life, including eukarya, archaea, and bacteria (Labunskyy et al. 2014). The selenoproteins mainly contain the glutathione peroxidase (GPx) family, iodothyronine deiodinase (DIO) family, thioredoxin reductases (TR) family, methionine-R-sulfoxide reductase 1 (MSRB1) family, thioredoxin-like (Rdx) family, etc. Even though they all contain Sec, the amount and location are different in these selenoproteins. Unquestionably, the UGA functions as a stop codon in the absence of a cis-acting stem-loop structure, designated Sec insertion sequence (SECIS) element in the 3′-UTR of selenoproteins (Walczak et al. 1998). SELENOW belongs to the Rdx family of selenoproteins and it plays an important role in antioxidation, responding to stress and involving cell immunity (Whanger 2009).
The low molecular weight selenoprotein SELENOW has a single Sec in its N-terminal region. It was initially discovered as a component absent in Se-deficient sheep with WMD (Vendeland et al. 1995). With the Sec residue at position 13 and the distinctive hallmark motif, Cys-X-X-Sec, the selenow isolated from a variety of animals contains 85–88 amino acids. (Whanger 2009). Moreover, SELENOW is located in the cytosol and the ubiquitous signaling adapter protein 14–3-3 is its target in mammals (Dikiy et al. 2007). Additionally, it has been reported that the Sec is required for SELENOW and 14–3-3 protein interactions (Jeon et al. 2016). Furthermore, according to a recent study, the protein SELENOW is downregulated in mice by RANKL/RANK/tumor necrosis factor receptor-associated factor 6/p38 signaling in response to receptor activator of nuclear factor (NF)-κB ligand (RANKL)-induced osteoclast differentiation (Kim et al. 2021).
Up to now, selenow gene has been identified in human, rat, mouse, monkey, pig, sheep, chicken (Li et al. 2011), zebrafish (Kryukov and Gladyshev 2000), pearl mussels (Hu et al. 2014), gold fish (Chen et al. 2015), topmouth culter (Dong et al. 2017) and yellow catfish (Xu et al. 2020). There were 3 selenow genes in zebrafish and 2 selenow genes in African clawed frog whereas just 1 selenow gene was found in yellow catfish and mammals. However, it is still unclear why the number of selenow gene was varied among different species. Although the selenow gene was expressed in a variety of tissues, including liver, spleen, kidney, muscle, skin, heart, intestine, etc., the tissue profiles were different among various species. In mammals, the highest expression levels of selenow mRNA were in the muscle, heart and brain (Whanger 2000). Nevertheless, the most abundant in mussels was in the hepatopancreas, much higher than in muscle (Chen et al. 2015; Hu et al. 2014). The expression of selenow mRNA was strongly regulated by Se concentrations, and Se deficiency could lead to a decrease in selenow mRNA expression (Chen et al. 2015; Liang et al. 2014; Sunde 2018; Yu et al. 2015).
Rainbow trout are one of the most popularly cultured cold-water commercial fish in the world. Wang et al. previously showed that the growth performance of rainbow trout was improved when fish were fed dietary supplementation with Se-yeast at dietary Se levels of 3.5–4.3 mg/kg, which is most closely related to the expression of the selenow gene (Wang et al. 2018). The hypothesis that a specific whole genome duplication (4R WGD) event occurred in the family ancestor of salmonid species between 50 and 100 million years ago is being supported by mounting evidence. This event is known to have caused duplicate copies of several important genes to be present in salmonid species (Guyomard et al. 2012; Marandel et al. 2019). Moreover, the average number of exons and introns significantly increased after the 4R WGD event in common carp (Lv et al. 2020). To the best of our knowledge, the characteristics of the selenow gene of rainbow trout have not been determined.
In our study, we cloned the partial cDNA sequence and detected the expression pattern of the rainbow trout selenow gene. The effect of dietary Se supplementation on the alterations of selenow expression was also studied. Our data provided a solid foundation for further investigating the biological function of selenoprotein in rainbow trout.
Materials and methods
All experiments involving animal care were conducted strictly according to the Guidance for the Care and Use of Laboratory Animals in China. This study was carried out under the guidelines of the Institutional Animal Care and Use Committees of Huazhong Agricultural University (Wuhan, China).
Fish maintenance and sampling
Healthy rainbow trout (mean body weight: 6.6 ± 1.0 g, mixed-sex) were kindly provided by Professor Zhen Xu from Huazhong Agriculture University (HZAU). Before gene cloning experiments, the fish were acclimatized to laboratory conditions in a circulating water tank by keeping the temperature at ~ 17 °C and were fed a commercial diet (Hubei Haid Feeds Company, Wuhan, China) twice a day (09:00 and 16:00).
The fish were sampled to clone the cDNA sequence of the selenow gene and examine its mRNA expression profile in various tissues. They were euthanized with tricaine methane sulfonate (MS-222, 100 mg/L) before dissection, and then the liver, spleen, kidney, head kidney, heart, brain, dorsal muscle, abdominal muscle, foregut, midgut, hindgut and gill were rapidly collected on ice. They were rapidly frozen in liquid nitrogen until RNA extraction.
RNA extraction and cDNA synthesis
Total RNA was extracted from different tissues using TRIzol Reagent (Invitrogen, USA) according to the manufacturer's protocol. The purity and concentration of total RNA were determined using a Nanodrop ND-2000 spectrophotometer (Thermo Electron Corporation, USA). The integrity of the total RNA was checked by 1.0% agarose gel electrophoresis. The first-strand cDNA was synthesized from 2 μg of total RNA by using the Revert Aid™ M-MLV Reverse Transcriptase Kit (Promega, USA) following the manufacturer's protocols. The cDNA products were stored at − 20 °C.
Molecular cloning of selenow gene
To amplify the core sequence of the selenow gene, different gene-specific primer pairs (selW-ORF-F1/R1) were designed based on the GenBank database (Table 1). The PCR reactions were performed in a total volume of 20 μL, including 10 μL premix Taq DNA polymerase (Yeasen, Shanghai, China), 2 μL (32 ng) cDNA, 0.8 μL (10 μM) of each primer and 6.4 μL ddH2O. The PCR thermal cycling programs were set as follows: initial denaturation at 95 °C for 3 min; then 35 cycles of 95 °C for 15 s, annealing temperature for 30 s and 72 °C for 40 s; 72 °C for 7 min and 16 °C for 5 min.
To gain the 3′-untranslated region (UTR) sequence of the selenow gene, we designed two gene-specific primers according to the core cDNA sequence of this gene and performed the 3′ rapid amplification of cDNA ends (RACE) PCR with the linker adapter (Table 1). In a first-round PCR reaction with first-strand cDNA as the template, the 3′ RACE Oligo (dT)17 primer and the SelW-outer primer were used. The 3′RACE Linker adapter and the SelW-inner primer were used in a second-round PCR reaction with the diluted product of the first-round PCR reaction as the template. The first-round PCR conditions were as follows: denaturation at 95 °C for 3 min; 31 cycles of denaturation at 94 °C for 30 s, annealing at 71 °C for 30 s, and extension at 72 °C for 45 s; 20 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s, and extension at 72 °C for 45 s; and a final extension at 72 °C for 10 min. The second-round PCR conditions were the same. All the primers were synthesized by the Tsingke Biotech Company (Wuhan, China). All PCR products were detected by 1.5% agarose gel electrophoresis, and products were purified using an AxyPrep™ gel extraction kit (Axygen, USA). 4.5 μL of purified PCR products were ligated into a 0.5 μL pMD18-T vector with another 5 μL Solution I (TaKaRa, Japan). Subsequently, 10 μL of ligated products were transformed into 100 μL competent Escherichia coli DH5@ cells (TaKaRa, Japan) at 16 °C for 3 h, and then mixed with 890 μL LB liquid medium and cultured at 37 °C for 1 h. Three selected positive colonies were checked by PCR and agarose gel electrophoresis and were sequenced by the Tsingke Biotech Company (Wuhan, China).
Quantitative real-time PCR (qPCR)
qPCR was used to detect mRNA expression levels of the selenow gene in various tissues of rainbow trout using a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, USA). The gene-specific primer pairs for qPCR were designed based on the cloned cDNA sequence of the selenow gene (Table 1). The EF1α and β-actin genes were used as internal control genes. The PCR reaction mixture consisted of 10 μL Hieff®qPCR®Green Master Mix (Yeasen, Shanghai, China), 4.4 μL ddH2O, 4 μL cDNA (10 times dilution of the template) and 0.8 μL of each gene-specific primer (10 μM) in a total volume of 20 μL. The qPCR of each sample was performed in triplicate according to the following conditions: 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s, 63 °C for 30 s and 72 °C for 30 s respectively. At the end of each PCR reaction, amplification curve and melting curve analyses were performed to check the integrity of the reaction and the quality of the product, respectively. To compare gene expression of selenow gene, the 2−∆∆Ct method was adopted to calculate the relative expression levels of the target gene (Livak and Schmittgen 2001).
Se diets
Healthy rainbow trout (mixed-sex) were purchased from the Enshi Guoxi Fishery Development Co. Ltd. (Hubei, China). Three diets—the Se-deficient diet (DSe), the Se-adequate diet (ASe) and the Se-excessive diet (ESe), were supplemented with Se-yeast (Angel Yeast Co., Ltd., Hubei, China, total Se content: 4 g/kg) at doses of 0, 4.0 and 16.0 mg/kg (Zhang et al. 2021). Dietary Se contents were measured by inductively coupled plasma-atomic emission spectrometry (ICP-MS) (Agilent 7500 c, Yokogawa Analytical Systems, Tokyo, Japan), and the actual Se contents in each diet were 0.09, 3.72 and 16.44 mg/kg, respectively. Water with a total Se content of 0.34 μg/L was detected. Fish were fed experimental diets for 10 weeks, and their muscle, spleen, gut, liver and brain were used for the determination of the mRNA level for the selenow.
Sequence analysis
The cDNA sequence and the deduced amino acid sequence were compared with the sequences in the GenBank database using the NCBI BLAST program. The theoretical isoelectric point and molecular weight of the amino acid were predicted using ExPASy (https://web.expasy.org/compute_pi/). The phosphorylation sites were predicted by using ExPASy (https://prosite.expasy.org/). The SECIS structure was predicted by using Seblastian (https://seblastian.crg.es/). Amino acid sequence similarity and identity were computed using the Sequence Manipulation Suite (http://www.bio-soft.net/sms/index.html). The ClustalW program in MEGA 6.06 and BoxShade (http://www.ch.embnet.org/software/BOX_form.html) were used for multiple sequence alignments. The phylogenetic tree of vertebrate SELENOW was constructed with Mega 6.06 by the neighbor-joining method, and the reliability was evaluated by the bootstrap method with 1000 replicates. The three-dimensional structure of SELENOW was established based on mouse SELENOW as the template using SWISS-MODEL (http://www.swissmodel.espasy.org).
One-way analysis of variance (ANOVA) test was performed on the data using SPSS 19.0 software (IBM, USA). The Duncan's multiple range test revealed that there were significant differences between the treatments. The difference was considered to be statistically significant if P < 0.05. All data were presented as the mean ± standard error of mean (SEM) (n = 3). All histograms were plotted using GraphPad Prism 8.0 software.
Result
Characterization of rainbow trout selenow cDNA and protein sequence
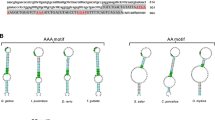
The partial cDNA sequence of the selenow gene was cloned from rainbow trout (Fig. 1). Selenoprotein W cDNA covered 765 nucleotides with an ORF of 393 nucleotides encoding 130 aa and a 3′-UTR of 372 nucleotides. The 3′-UTR contains one mRNA unstable motif (ATTTA). There are three protein kinase c phosphorylation sites in the SELENOW at the amino acid residue 7 (SLR), 35 (SRR) and 103 (SKK), together with three casein kinase II phosphorylation sites at the amino acid residue 68 (TQLE), 103 (SKKE) and 119 (TLVD).
Nucleotides and deduced amino acid sequences of the selenow cDNA from rainbow trout. The start codon (ATG) and stop codon (TAA) of open reading frame are marked as gray shadow and the TGA codon is in bold and selenocysteine (TGA, U) are boxed. Lowercase letters indicate the 3′-untranslated region (UTR), where the predicted selenocysteine insertion sequence (SECIS) is indicated by dashed line. The mRNA instability motif (ATTTA) is underlined with the broken line. The phosphorylation sites are underlined with thin lines under amino acid sequences. The GenBank accession number of this gene is listed in Table 2
The amino acid in SELENOW was estimated to possess a theoretical isoelectric point (PI) and molecular weight (MW) of 9.01 and 14.713 kDa, respectively. The TGA (terminal codon) codes for the Sec in the 57th aa residue of SELENOW.
SECIS element analysis of selenow
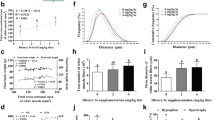
One SECIS element was discovered in the 3′-UTR of selenow using SECISearch3, resulting in a stem-loop secondary structure of the mRNA sequence (Fig. 2). The SECIS element of selenow was 81 bp. The type I SECIS element, which is found in the selenow mRNA, contains two helixes, an apical loop, one internal loop and a quartet of non-Watson–Crick tandemly linked bases (GA quartet). The GA quartet, which includes the conservative sequences UGAN and GA at the 5′ and 3′ bases of the stem, was found under 10 bp helix II and was made up of 4 non-Watson–Crick interacting base pairs. Under the SECIS core, the internal loop and helix I were found.
The predicted three-dimensional structure of rainbow trout SELENOW
The predicted 3D molecular modeling of rainbow trout SELENOW was shown in Fig. 3. Based on the secondary structure analysis, the rainbow trout SELENOW 3D model contained two α-helices, four β-sheets, and two loops. The amino-terminal to carboxy-terminal structure of the rainbow trout SELENOW followed a pattern of β1-α1-β2-β3-β4-α2.
Homology and phylogenetic analysis
The rainbow trout SELENOW shared many conserved or identical regions with other known vertebrates, according to multiple sequence alignment (Fig. 4). The amino acid sequence of SELENOW contained a conserved motif of 54CXXU57. According to the predicted amino acid sequence comparisons, the rainbow trout SELENOW had the highest similarity (94.6%) with the SELENOW of Atlantic salmon (Salmo salar), followed by 53.5% of similarity with common carp (Cyprinus carpio) and zebrafish (Danio rerio) (Table 3).
Multiple alignment of the deduced rainbow trout SELENOW amino acid sequences with those of other vertebrates, derived using the ClustalW program. The identical residues are indicated in black and marked with an asterisk (*). Conserved and semi-conserved residues are indicated by gray shading, marked with a dot (.). Absent amino acids are indicated by dashes (–)
The rainbow trout SELENOW was initially grouped with the SELENOW of Atlantic salmon, and later these two species were grouped with the four Cyprinidae fishes into a branch according to the NJ phylogenetic tree of deduced SELENOW amino acid sequences. This branch of fish was totally distinguishable from the mammal, bird and amphibian groups (Fig. 5).
Expression pattern analyses of selenow gene in different tissues
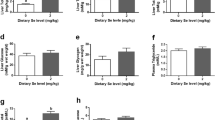
The distribution of selenow expression among various rainbow trout tissues was quantified using qPCR (Fig. 6). Results showed that selenow transcripts were broadly expressed in thirteen examined tissues at different levels. The dorsal muscle exhibited the highest level of expression, followed by the abdominal muscle and brain. The expression of selenow transcripts was low in the spleen, kidney, head kidney, heart, gill, liver and gut.
Tissue distribution of relative expression levels of selenow mRNAs in rainbow trout. qPCR was used to investigate the distribution of the selenow expression in different tissues from rainbow trout at the mRNA level. EF1α and β-actin were used as internal control genes for the relative quantification of cDNA in PCR reactions. The data were expressed as the mean fold change (mean ± SEM, n = 3) relative to the foregut. Bars that share different letters indicate significant differences among various tissues (P < 0.05)
Transcription response of selenow to Se levels in diets
To investigate the response of the selenow to Se levels in diets, rainbow trout were fed with three diets: the Se-deficient diet (DSe, 0 mg/kg), the Se-adequate diet (ASe, 4 mg/kg), and the Se-excessive diet (ESe, 16 mg/kg), respectively. After 10 weeks, we detected the total Se content in the muscle to confirm the effect of Se-supplementation. The results indicated that the Se-supplement increased the Se content in muscle effectively (Fig. S1). Next, we detected the expression levels of selenow mRNA in the muscle, brain, spleen, liver and gut. The expression level of selenow mRNA of the DSe group showed a declining tendency in comparison to the ASe group among all five investigated tissues. Interestingly, the mRNA level of the selenow gene was significantly decreased only in the muscle (P < 0.01). However, the ASe group and the Ese group did not significantly differ in terms of selenow expression level (Fig. 7). These results revealed that the selenow gene could respond to Se levels in diets and was sensitive in muscle.
Transcriptional response of selenow in muscle, spleen, liver, gut and brain of rainbow trout fed to three Se levels of diets. The reference genes are EF1α and β-actin. The data were expressed as the mean fold change (mean ± SEM, n = 3) relative to the ASe group. Values with significant differences compared to the ASe group are indicated by letters (P < 0.05)
Discussion
Understanding the structure of selenoproteins is crucial to comprehending their biological functions. In the current study, we identified the cDNA sequence of selenow from rainbow trout. The determined amino acid sequence of SELENOW from rainbow trout and the selected animals both shared the 54CXXU57 motif, which suggests that the function of SELENOW in rainbow trout may be similar to that of SELENOW in other vertebrates. However, fish, chicken and amphibians did not include the conserved amino acid Cys 37th, which is found in mammals' SELENOW, and the Cys 37th was substituted by threonine, arginine or aspartic acid, respectively. It was thought that the amino acid Cys 37th in mammals was involved in the binding of glutathione and SELENOW (Beilstein et al. 1996; Dong et al. 2017; Gu et al. 1999). Further investigation is necessary to determine whether the loss of the SELENOW residue Cys 37th would result in any variations between non-mammals and mammals. It's interesting to note that the SELENOW of rainbow trout and Atlantic salmon is longer than that of other vertebrates. It is likely due to the 4R WGD event that occurred in the salmonid, similar to the cyprinid (Guyomard et al. 2012).
The stem-loop secondary structure, which is found in the 3′-UTR of all eukaryotic and archaeal selenoprotein mRNAs, was predicted to be present in the rainbow trout SELENOW mRNA (Aachmann et al. 2007; Chambers et al. 1986; Labunskyy et al. 2014; Low and Berry 1996). Additionally, the SECIS component was necessary for translating UGA Sec codons (Walczak et al. 1998). The stem-loop secondary structure of rainbow trout SELENOW mRNA contains two helixes, an apical loop, an internal loop and a GA quartet, which is similar to the SECIS structure of SELENOW in fish and birds. Some conservative domains were crucial for the SECIS components of rainbow trout SELENOW. The core of SECIS was the GA quartet. Kink-turn motifs were distinguished by the tandem of G.A/A.G base pairs (Dong et al. 2017; Goody et al. 2004; Matsumura et al. 2003). The GA quartet is the primary component of SECIS so that it can interact with SECIS binding protein 2 (SBP2). According to RNA-footprinting studies, SBP2 predominantly binds to the GA quartet, the 5′-strand of the internal loop and the top half of helix I (Fletcher et al. 2001). In addition, it was necessary for Sec incorporation to get the conservative AAR motif in the apical loop (Martin et al. 1998). Depending on whether an additional loop is present or absent, eukaryotic SECIS elements are typically categorized as forms I or II, respectively (Chapple et al. 2009). Besides, the top of the SECIS element lacks an additional tiny stem-loop. As a result, the SECIS of rainbow trout SELENOW belonged to the SECIS I form. In contrast to mammals, it was similar to the SECIS structures of fish, shellfish and birds (Xu et al. 2020). However, further investigation is needed to determine the function of the additional tiny stem-loop.
Many researchers have found that the selenow gene displays different expression patterns in a variety of tissues, including muscle, spleen, liver and brain (Chen et al. 2015; Dong et al. 2017; Gu et al. 1999; Hu et al. 2014; Li et al. 2011; Xu et al. 2020). In humans, selenow gene expression was high in the muscle and heart, while in chickens, selenow was the predominant expression in nervous tissue, muscle, gizzard, blood vessel and cartilaginous tissue (Li et al. 2011). In the yellow catfish, the expression of the selenow gene was high in mesenteric fat, gill, spleen and muscle (Xu et al. 2020). Similarly, pearl mussel selenow gene mRNA levels were high in the hepatopancreas and weak in the hemocytes (Hu et al. 2014). In this study, the mRNA of selenow in rainbow trout was extensively distributed in all selected tissues, including muscle, brain, spleen, kidney, head kidney, heart, gill, liver and gut. The wide range of expression of selenow in rainbow trout is consistent with the previous research. Additionally, the widespread expression of the selenow gene implicates its multiplicity of biological functions.
It has been known that selenow expression is modulated by Se levels in rats, mice, sheep and rainbow trout (Wang et al. 2018; Yeh et al. 1998). Previous study has shown that selenow level increases with dietary Se supplementation (Yu et al. 2011). In the present study, the transcription response of selenow to dietary Se levels was evaluated in the muscle, spleen, liver, brain and gut. With the increase of dietary Se level, the mRNA level of selenow gene was significantly increased only in the muscle (P < 0.01). We suspect that the selenow gene might be more sensitive to Se treatment in muscle compared to other tissues. However, Se deficiency reduced the mRNA levels of the selenow in all detected tissues. Excessive Se increased the selenow expression in the spleen and liver, but not in the gut and brain. However, the mRNA level of the selenow gene was increased in low and high Se concentrations in the spleen and brain in yellow catfish. Whereas there was no significance in the mRNA level of selenow in the kidney (Xu et al. 2020). The discrepancy of selenow gene response to Se levels between the yellow catfish and rainbow trout might be due to the different Se forms. Together with these results, the expression profiles in response to Se levels in various tissues suggest that selenow may play a vital role in the muscle. There is a major limitation in this study that could be addressed in further research. This study was based on the mRNA levels but not protein levels. The role of SELENOW needs further investigation at the protein level.
In conclusion, a novel selenow cDNA was cloned and characterized from rainbow trout. All analysis of the sequence, structures and evolution indicates the selenow belongs to the selenoprotein family. The gene has been widely expressed in rainbow trout tissues. Furthermore, our research highlights the vital role of selenow in rainbow trout muscle response to dietary Se levels.
References
Aachmann FL, Fomenko DE, Soragni A, Gladyshev VN, Dikiy A (2007) Solution structure of selenoprotein W and NMR analysis of its interaction with 14–3-3 proteins. J Biol Chem 282:37036–37044. https://doi.org/10.1074/jbc.M705410200
Beilstein MA, Vendeland SC, Barofsky E, Jensen ON, Whanger PD (1996) Selenoprotein W of rat muscle binds glutathione and an unknown small molecular weight moiety. J Inorg Biochem 61:117–124. https://doi.org/10.1016/0162-0134(95)00045-3
Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, Harrison PR (1986) The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the ‘termination’ codon, TGA. EMBO J 5:1221–1227. https://doi.org/10.1002/j.1460-2075.1986.tb04350.x
Chapple CE, Guigó R, Krol A (2009) SECISaln, a web-based tool for the creation of structure-based alignments of eukaryotic SECIS elements. Bioinformatics 25:674–675. https://doi.org/10.1093/bioinformatics/btp020
Chen W, Zhang Z, Dong H, Jiang X (2015) Molecular cloning and sequence analysis of selenoprotein W gene and its mRNA expression patterns in response to metabolic status and cadmium exposure in goldfish, Carassius auratus. Comp Biochem Physiol B 184:1–9. https://doi.org/10.1016/j.cbpb.2015.01.005
Dikiy A, Novoselov SV, Fomenko DE, Sengupta A, Carlson BA, Cerny RL, Ginalski K, Grishin NV, Hatfield DL, Gladyshev VN (2007) SelT, SelW, SelH, and Rdx12: genomics and molecular insights into the functions of selenoproteins of a novel thioredoxin-like family. Biochemistry 46:6871–6882. https://doi.org/10.1021/bi602462q
Dong H, Chen W, Sun C, Sun J, Wang Y, Xie C, Fu Q, Zhu J, Ye J (2017) Identification, characterization of selenoprotein W and its mRNA expression patterns in response to somatostatin 14, cysteamine hydrochloride, 17β-estradiol and a binary mixture of 17β-estradiol and cysteamine hydrochloride in topmouth culter (Erythroculter ilishaeformis). Fish Physiol Biochem 43:115–126. https://doi.org/10.1007/s10695-016-0272-9
Fletcher JE, Copeland PR, Driscoll DM, Krol A (2001) The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA 7:1442–1453
Gladyshev VN (2001) Identity, evolution and function of selenoproteins and selenoprotein genes. In: Hatfield DL (ed) Selenium. Springer, Boston
Goody TA, Melcher SE, Norman DG, Lilley DMJ (2004) The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA 10:254–264. https://doi.org/10.1261/rna.5176604
Gu QP, Beilstein MA, Barofsky E, Ream W, Whanger PD (1999) Purification, characterization, and glutathione binding to selenoprotein W from monkey muscle. Arch Biochem Biophys 361:25–33. https://doi.org/10.1006/abbi.1998.0949
Guyomard R, Boussaha M, Krieg F, Hervet C, Quillet E (2012) A synthetic rainbow trout linkage map provides new insights into the salmonid whole genome duplication and the conservation of synteny among teleosts. BMC Genet 13:15. https://doi.org/10.1186/1471-2156-13-15
Hoffmann PR, Berry MJ (2005) Selenoprotein synthesis: a unique translational mechanism used by a diverse family of proteins. Thyroid 15:769–775. https://doi.org/10.1089/thy.2005.15.769
Hu B, Liu Y, Wen C-G, Li A-H, Hu X-P, Wu D, Hu X-J, Tao Z-Y (2014) Cloning and expression of selenoprotein W from pearl mussels Cristaria plicata. Comp Biochem Physiol B 167:8–15. https://doi.org/10.1016/j.cbpb.2013.09.008
Jeon YH, Ko KY, Lee JH, Park KJ, Jang JK, Kim IY (2016) Identification of a redox-modulatory interaction between selenoprotein W and 14–3-3 protein. Biochim Biophys Acta 1863:10–18. https://doi.org/10.1016/j.bbamcr.2015.10.006
Kim H, Lee K, Kim JM, Kim MY, Kim JR, Lee HW, Chung YW, Shin HI, Kim T, Park ES, Rho J, Lee SH, Kim N, Lee SY, Choi Y, Jeong D (2021) Selenoprotein W ensures physiological bone remodeling by preventing hyperactivity of osteoclasts. Nat Commun 12:2258. https://doi.org/10.1038/s41467-021-22565-7
Kryukov G, Gladyshev V (2000) Selenium metabolism in zebrafish: multiplicity of selenoprotein genes and expression of a protein containing 17 selenocysteine residues. Genes Cells 5:1049–1060. https://doi.org/10.1046/j.1365-2443.2000.00392.x
Labunskyy VM, Hatfield DL, Gladyshev VN (2014) Selenoproteins: molecular pathways and physiological roles. Physiol Rev 94:739–777. https://doi.org/10.1152/physrev.00039.2013
Li JL, Ruan H, Li HX, Li S, Xu SW, Tang ZX (2011) Molecular cloning, characterization and mRNA expression analysis of a novel selenoprotein: avian selenoprotein W from chicken. Mol Biol Rep 38:4015–4022. https://doi.org/10.1007/s11033-010-0520-5
Liang Y, Lin S, Wang C, Yao H, Zhang Z, Xu SW (2014) Effect of selenium on selenoprotein expression in the adipose tissue of chickens. Biol Trace Elem Res 160:41–48. https://doi.org/10.1007/s12011-014-0024-6
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Low SC, Berry MJ (1996) Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci 21:203–208. https://doi.org/10.1016/S0968-0004(96)80016-8
Lv H, Zhou T, Dong C, Kong S, Chen L, Pu F, Li X, Xu P (2020) Genome-wide identification, evolution, and mRNA expression of complement genes in common carp (Cyprinus carpio). Fish Shellfish Immunol 96:190–200. https://doi.org/10.1016/j.fsi.2019.11.032
Marandel L, Kostyniuk DJ, Best C, Forbes JLI, Liu J, Panserat S, Mennigen JA (2019) Pck-ing up steam: widening the salmonid gluconeogenic gene duplication trail. Gene 698:129–140. https://doi.org/10.1016/j.gene.2019.02.079
Martin GW, Harney JW, Berry MJ (1998) Functionality of mutations at conserved nucleotides in eukaryotic SECIS elements is determined by the identity of a single nonconserved nucleotide. RNA 4:65–73
Matsumura S, Ikawa Y, Inoue T (2003) Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res 31:5544–5551. https://doi.org/10.1093/nar/gkg760
Rayman MP (2000) The importance of selenium to human health. Lancet 356(9225):233–241. https://doi.org/10.1016/S0140-6736(00)02490-9
Sunde RA (2018) Selenium regulation of selenoprotein enzyme activity and transcripts in a pilot study with founder strains from the collaborative cross. PLoS ONE 13:e0191449. https://doi.org/10.1371/journal.pone.0191449
Vendeland SC, Beilstein MA, Yeh JY, Ream W, Whanger PD (1995) Rat skeletal muscle selenoprotein W: cDNA clone and mRNA modulation by dietary selenium. Proc Natl Acad Sci USA 92:8749–8753. https://doi.org/10.1073/pnas.92.19.8749
Walczak R, Carbon P, Krol A (1998) An essential non-Watson-Crick base pair motif in 3’UTR to mediate selenoprotein translation. RNA 4:74–84
Wang L, Zhang XZ, Wu L, Liu Q, Zhang DF, Yin JJ (2018) Expression of selenoprotein genes in muscle is crucial for the growth of rainbow trout (Oncorhynchus mykiss) fed diets supplemented with selenium yeast. Aquaculture 492:82–90. https://doi.org/10.1016/j.aquaculture.2018.03.054
Whanger PD (2000) Selenoprotein W: a review. CMLS. Cell Mol Life Sci 57:1846–1852. https://doi.org/10.1007/pl00000666
Whanger PD (2009) Selenoprotein expression and function-selenoprotein W. Biochim Biophys Acta 1790:1448–1452. https://doi.org/10.1016/j.bbagen.2009.05.010
Xu X, Zhang DG, Zhao T, Xu YH, Luo Z (2020) Characterization and expression analysis of seven selenoprotein genes in yellow catfish Pelteobagrus fulvidraco to dietary selenium levels. J Trace Elem Med Biol 62:126600. https://doi.org/10.1016/j.jtemb.2020.126600
Yeh JY, Ou BR, Gu QP, Whanger P (1998) Influence of gender on selenoprotein w, glutathione peroxidase and selenium in tissues of rats*. Comp Biochem Physiol B 119:151–155. https://doi.org/10.1016/S0305-0491(97)00298-8
Yu D, Li JL, Zhang J, Gao X, Xu SW (2011) Effects of dietary selenium on selenoprotein W gene expression in the chicken immune organs. Biol Trace Elem Res 144:678–687. https://doi.org/10.1007/s12011-011-9062-5
Yu D, Zhang Z, Yao H, Li S, Xu SW (2015) The role of selenoprotein W in inflammatory injury in chicken immune tissues and cultured splenic lymphocyte. Biometals 28:75–87. https://doi.org/10.1007/s10534-014-9804-x
Zhang F, Teng ZL, Wang L, Wang L, Huang TT, Zhang XZ (2021) Dietary selenium deficiency and excess accelerate ubiquitin-mediated protein degradation in the muscle of rainbow trout (Oncorhynchus mykiss) via Akt/FoxO3a and NF-κB Signaling pathways. Biol Trace Elem Res. https://doi.org/10.1007/s12011-021-02726-x
Acknowledgements
This work was financially supported by the National Key R&D Program (Grant Number: 2019YFD0900303) and the Fundamental Research Funds for the Central Universities (Grant Numbers: 2662021SPPY001 and 11900022414).
Author information
Authors and Affiliations
Contributions
CL: conceptualization, investigation, formal analysis, writing-original draft. FZ, ZT and GZ: methodology, formal analysis, writing—review & editing. YY, PX, XH, LW, FY and ZY: methodology, formal analysis. XZ: writing-review & editing, supervision, project administration, funding acquisition. All the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liao, C., Zhang, F., Teng, Z. et al. Molecular characterization and expression analysis of selenoprotein W gene in rainbow trout (Oncorhynchus mykiss) with dietary selenium levels. Biometals 35, 1359–1370 (2022). https://doi.org/10.1007/s10534-022-00451-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-022-00451-z