Abstract
Iron is reported to interact with other metals. In addition, it has been shown that genetic background may impact iron metabolism. Our objective was to characterize, in mice of three genetic backgrounds, the links between iron and several non-iron metals. Thirty normal mice (C57BL/6, Balb/c and DBA/2; n = 10 for each group), fed with the same diet, were studied. Quantification of iron, zinc, cobalt, copper, manganese, magnesium and rubidium was performed by ICP/MS in plasma, erythrocytes, liver and spleen. Transferrin saturation was determined. Hepatic hepcidin1 mRNA level was evaluated by quantitative RT-PCR. As previously reported, iron parameters were modulated by genetic background with significantly higher values for plasma iron parameters and liver iron concentration in DBA/2 and Balb/c strains. Hepatic hepcidin1 mRNA level was lower in DBA/2 mice. No iron parameter was correlated with hepcidin1 mRNA levels. Principal component analysis of the data obtained for non-iron metals indicated that metals parameters stratified the mice according to their genetic background. Plasma and tissue metals parameters that are dependent or independent of genetic background were identified. Moreover, relationships were found between plasma and tissue content of iron and some other metals parameters. Our data: (i) confirms the impact of the genetic background on iron parameters, (ii) shows that genetic background may also play a role in the metabolism of non-iron metals, (iii) identifies links between iron and other metals parameters which may have implications in the understanding and, potentially, the modulation of iron metabolism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Iron metabolism is finely tuned in order to ensure health and wellbeing (Andrews 1999; Pollack et al. 1965). Disorders of iron metabolism include genetic iron overload, especially those related to the p.Cys282Tyr in the HFE gene. Bioclinical findings in HFE hemochromatosis show that the phenotypic presentation of the disease is variable. Indeed, whereas three per thousand persons exhibit homozygous mutation of HFE gene, and are therefore genetically predisposed, the disease expression spectrum may vary from the absence of iron metabolism abnormalities to the sole presence of transferrin saturation increase reflecting an abnormal recycling of iron, and to the development of visceral iron-related complications with an impact on both quality of life and life expectancy (Brissot et al. 2008). Similarly, chronic inflammation occurring in many diseases may inconstantly impact iron metabolism (Weiss and Goodnough 2005), exposing patients to the risk of anaemia of chronic disease.
In order to understand such differences, studies have been conducted to identify iron genes which could participate to this variability. Thus, other genes involved in iron metabolism have been identified, when mutated, in non-HFE genetic hemochromatosis (Camaschella et al. 2000; Papanikolaou et al. 2004; Roetto et al. 2003). In addition, mutations within these genes could play a role in the expression of HFE hemochromatosis (Island et al. 2009; Le Gac et al. 2004; Merryweather-Clarke et al. 2003; Milet et al. 2007) or other iron related diseases (An et al. 2012; Finberg et al. 2008; Finberg et al. 2011; Melis et al. 2008; Nai et al. 2011). The role of polymorphisms in non-iron genes has also been evoked. In addition, differences of clinical expression between genders could be related to testosterone (Latour et al. 2014). In mice, regarding genetic background, variability of iron gene expression has been reported between different strains (Courselaud et al. 2004; Dupic et al. 2002). In addition, iron overload severity in Hfe −/− mice is modulated by genetic background (Coppin et al. 2007). However, to date, there is no clear explanation for such a variability in the phenotypic expression of iron-related diseases in humans.
The observation of iron-related diseases in patients with gene mutations associated to other metals raises the general issue of the impact of non-iron metals on iron metabolism. Thus, iron overload is observed in hereditary aceruloplasminemia (Harris et al. 1995; Loreal et al. 2002), but also in acquired hypo-ceruloplasminemia associated with zinc exposure (Videt-Gibou et al. 2009). In both situations, the lack of ferroxidase activity of ceruloplasmin, which is dependent of copper, likely alters iron transport in plasma. Moreover, abnormalities of iron metabolism have been reported in presence of metabolic alterations related to non-iron metals such as cobalt and manganese (Garcia et al. 2007; Hansen et al. 2010; Simonsen et al. 2012).
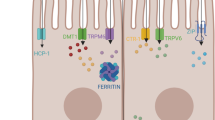
The potential interaction between iron and non-iron metals is also emerging from the observation that DMT1 (SLC11A2 gene), which is involved in the crossing of iron toward membranes, was also reported to take in charge other divalent cations including: zinc (Zn2+), copper (Cu2+), nickel (Ni2+), cobalt (Co2+), cadmium (Cd2+), manganese (Mn2+) and lead (Pb2+) (Gunshin et al. 1997). Moreover, ferroportin, the exporter of iron from enterocytes and macrophages toward the plasma, could also export zinc and cobalt (Mitchell et al. 2014; Troadec et al. 2010). It has also been reported that the expression of hepcidin, the main controller of systemic iron metabolism, could be modulated by the metal responsive element- binding transcription factor-1 (MTF1) (Balesaria et al. 2010).
Taken together, these elements suggest that interactions with non-iron metals may play a role in the control of normal, and abnormal, iron metabolism. Therefore, our goal was, by taking advantage of reported differences on iron metabolism between mice from different genetic backgrounds, to look whether other metals are also modulated by genetic background and to search for relationships between hepcidin, iron and non-iron metals. The study was conducted in DBA/2, C57BL/6, Balb/c wild-type mouse strains, and we characterized 6 metals, including: (i) four transition metals Zn, Cu, Mn, Co which have been associated with iron metabolism in the literature; (ii) Mg, an earth alkali metal cofactor of numerous enzymes (Fawcett et al. 1999) and (iii) Rb, an alkali metal, reported as a trace element having biological impact on behaviour (Carroll and Sharp 1971) and spermatogenesis (Yamaguchi et al. 2007), and being modulated by genetic background in mice (Siegers et al. 1977). Our data supports an impact of genetic background on the metabolism of non-ferrous metals, in addition to iron metabolism, and identifies links between iron and other metals parameters, related or not to genetic background.
Materials and methods
Animals
The study was approved by the ethical Rennes committee for animal experimentation under the reference R-2012-0L-02. Four week-old C57BL/6 J, Balb/cJ and DBA/2 J mice were obtained from the Centre d’Elevage Robert Janvier (Le Genest St Isle, France) and housed in animal facilities (ARCHE) of UMS Biosit in Rennes. Thirty male mice (ten per group) were included. The environment was temperature and light-controlled. Mice were given free access to water and food (standard diet CRM-E from Special Diet Services®) over 4 weeks. All mice were studied at 8 weeks, in the morning between 9 and 12 am. Blood was obtained from a trans-diaphragmatic intracardiac puncture, and sampled in sodium heparin tubes suitable for trace element analysis. Mice were sacrificed and livers and spleens were dissected and weighed. Samples of liver and spleen were quickly frozen in liquid nitrogen, and then stored at −80 °C to perform the measurement of trace element concentrations and mRNA extractions. In addition, liver and spleen samples were also fixed in 4 % buffered-formaldehyde for histological studies.
Histological analysis
After formaldehyde fixation, samples were paraffin embedded, and tissue sections (5 µm) were performed. For histological evaluation of iron deposits, Perl’s staining and light microscopy evaluation were performed.
Haemoglobin level determination
Haemoglobin level was determined during the sacrifice on blood sample using HemoCue® 201+.
Plasma iron parameters and plasma magnesium quantification
Plasma iron and unsaturated iron-binding capacity (UIBC) were measured in the biochemistry laboratory (Rennes Hospital) on Cobas 8000 analyzer Roche® (Cobas® reagents 03183696 122 and 04536355 190 respectively). Plasma transferrin saturation was then calculated as (plasma iron/(plasma iron + UIBC)) X100.
Plasma magnesium was measured on Cobas 8000 analyzer Roche® (Cobas® 20737593 322).
Trace elements
Sample preparation: plasma, erythrocytes, liver and spleen
All samples were handled with special care in order to avoid environmental contamination.
After blood sample centrifugation, plasma was taken and frozen in polypropylene cryotubes at −80 °C. Erythrocytes were then washed three times in NaCl 0.9 %, centrifuged in NaCl 0.9 % (3000 g/min), and then were frozen in polypropylene cryotubes at −80 °C.
Livers and spleens samples were desiccated for 15 h at 120 °C. Then, dried samples were weighed and mineralized by nitric acid solution (Fisher Chemical—Optima Grade®) in special polypropylene tubes for 2 h at 110 °C in a heating block. Specimens were preserved at 4 °C until quantification of metals.
Trace elements quantification
Studied elements were: magnesium (24Mg), manganese (55Mn), iron (56Fe), cobalt (59Co), copper (63Cu), zinc (66Zn), and rubidium (85Rb). Trace elements were measured by ICP-MS (Inductively Coupled Plasma Mass Spectrometry), on a X-Series II from Thermo Scientific® equipped with collision cell technology (Platform AEM2, University of Rennes 1/Biochemistry Laboratory, University Rennes Hospital). The source of plasma was argon (Messer®) with high degree of purity (>99.999 %). The collision/reaction cell used was pressurized with a mixture of helium (93 %) and hydrogen (7 %) (Messer®). Ultrapure water was obtained from Millipore Direct-Q® 3 water station. Nitric acid solution was suprapur, at 69 % (Fisher Chemical—Optima Grade®). Surfactant agent was used (Triton X-100—Merck®). The internal standard used was rhodium (Fisher Scientific®). Calibration ranges preparation was carried out using a multi-element calibrator solution (SCP Science® Plasma Cal). Calibration and verification of instrument performance were realized using multi-element solutions, respectively tune F and tune A (Thermo®). Certified reference materials and quality control were respectively obtained from NCS (bovine liver ZC71001) and Recipe (Clincheck plasma and whole blood controls for trace elements).
Quantification of hepcidin 1 mRNA
Expression of hepatic specific transcripts was analysed by quantitative RT-PCR. Total RNA from mouse liver was isolated using the SV Total RNA Isolation System (Promega®). RNA was then reverse transcribed into cDNA with the M-MLV reverse transcriptase (Promega®). The following primers have been used to amplify hepcidin 1 (Hepc1) (forward: 5′-TTCCCAGTGTGGTATCTGTTGC-3′ and reverse: 5′-GGTCAGGATGTGGCTCTAGGC-3′), and HPRT (forward: 5′-TGTTCTAGTCCTGTGGCCATCT3′ and reverse: 5′-GCTCAT AGTGCAAATCAAAAGTCT-3′). Quantitative real-time PCR assays were performed in 96-well microplates using the qPCR MasterMix Plus for SYBR® Green I (Eurogentec®) and the system StepOne Plus (Real-Time PCR System—Applied Biosystems®). All results were analysed by StepOne Software v2.1 (Applied Biosystem®). For each cDNA sample, the difference between the threshold cycle for Hepc1 amplification and the threshold cycle for HPRT was calculated. This allowed normalization of the amount of target to the endogenous reference, HPRT.
Statistical analysis
Results were expressed as mean ± SD. Results obtained in the 3 strains were firstly compared by nonparametric Kruskall-Wallis test, followed when appropriate by a pair-wise comparison using nonparametric Mann–Whitney test. A p < 0.05 was considered significant.
In order to get an overview of possible associations that could exist in different murine genetic backgrounds for trace elements and iron parameters, the results were explored by principal component analysis (PCA). Analyses were performed with Factor MineR library (http://cran.r-project.org). PCA aims to simplify a dataset by reducing their dimensionality, while maintaining the maximum of information contained in these data. Thus, PCA provides graphical representations from the maximum of information in a data table. It allows the examination of the correlations between the variables (here the determined parameters) and to prioritize them according to their link to the principal components (PCs) (here the two PCs that capture maximum inertia highlight difference in the genetic backgrounds).
Associations between parameters were assessed by univariate linear regression analysis. When the strain or the body weight was significantly associated with a parameter of interest, a multivariate linear regression analysis, including these variables, was performed.
Results
Genetic background influences iron metabolism and non-iron metals
Quantifications of trace elements—Co, Mn, Zn, Cu, Rb, Mg and Fe—were performed in plasma, liver, spleen and erythrocytes. In addition, plasma transferrin saturation, liver hepcidin 1 mRNA and haemoglobin levels were determined. Values obtained are summarized in Table 1 and schematic representation of the significant differences between the three strains is presented in Supplemental data S1.
Briefly, regarding the impact of genetic background on each metal concentration: (i) for Co, values were significantly different between strains in liver and erythrocytes; (ii) for Mn and Zn, significant differences appeared only in erythrocytes; (iii) for Cu, significant differences existed in plasma, liver and erythrocytes; (iv) it is noteworthy that, for Rb, significant differences were found in the four studied matrices; (v) for Mg, values in plasma, liver and spleen significantly differed between strains; (vi) for Fe, significant differences were found in plasma and liver; in addition, transferrin saturation and hepcidin 1 mRNA quantification were affected according to strains.
Regarding the impact of strain on metal quantification in the different tissues: (i) in erythrocytes, all metals, but Fe and Mg, were modulated; (ii) in the liver, all metals, but manganese and zinc, were affected; (iii) in plasma, Cu, Rb, Mg and Fe were modulated; (iv) in the spleen, only Rb and Mg were modulated.
Global analysis of the impact of genetic background on metals
We performed PCA to explore relationships existing between genetic backgrounds and metals concentrations, and particularly whether or not the three strains differed regarding the various studied biochemical parameters.
The first principal component (PC 1) explains 22.7 % of parameters variability. PC 1 primarily identifies differences between C57BL/6 mice and DBA/2 mice, but also reveals a stratification of all genetic backgrounds (Fig. 1a). PC 2 likely represents inter-individual variations. Vectors representing the most influential biochemical parameters which explain differences between strains, are presented in Fig. 1b. Iron parameters, including plasma iron, transferrin saturation and spleen iron are involved in the stratification. The Rb levels in the four studied matrices (plasma, liver, spleen and erythrocytes) play also an important role in the stratification. Cobalt and copper, mainly erythrocytes, plasma and liver concentrations, are also involved, as well as the spleen and erythrocytes zinc and manganese concentrations. Variables playing a major role in the stratification according to axis 1 in PCA analysis are found in supplemental data S2.
PCA analysis studying associations in different murine genetic backgrounds between trace elements and iron related parameters. A Mapping of mice regarding the first two-dimensions (axis 1 and 2). Each mouse is represented by a dot: Balb/c mice in black, C57BL/6 mice in red and DBA/2 mice in green. The open squares represent the geometrical centroids for each strain. B Representation of variables playing a major role in the stratification according to axis 1
Links between metal parameters
Firstly, we identified by univariate linear regression analysis the metal parameters associated with a strain effect (Table 2). Thereafter, in each matrix, associations between all trace elements were studied through linear regression models. When univariate linear regression analysis led us to identify association between a parameter and the strain (Table 2), a multivariate linear regression including the strain was performed. Results are schematically presented in Fig. 2.
Schematic representation of the existing links between trace elements and iron parameters values in the four matrices. Relationships were studied for each element in each matrix. They were determined by univariate linear regression analysis, when the strain was demonstrated having no impact on the parameter, or by multivariate linear regression analysis, including strain, when an impact of genetic background was demonstrated. Parameters, for which a strain effect was determined, are indicated by asterisk. Associations with a p value < 0.01 are indicated by a thick line, associations with a p value between 0.05 and 0.01 by a thin line. Associations identified by multivariate linear regression analysis (p value < 0.05) but not found by univariate analysis, are indicated as dotted lines. Liver hepcidin 1 mRNA level, considered as a potential regulator of trace elements, has been integrated and taken into account in the four matrices
In plasma, associations were observed particularly between liver hepcidin 1 mRNA and Mn, and between transferrin saturation and copper. There was no relationship between plasma iron or transferrin saturation and hepcidin 1 mRNA level. In erythrocytes, an association was observed between liver hepcidin 1 mRNA and haemoglobin levels. Moreover, a large number of associations were observed between the different trace elements. Finally, in liver and spleen, associations were noted mainly between Fe, Co and Mn, as well as between liver hepcidin 1 mRNA and Rb.
Discussion
The variable phenotypic features of iron-related diseases, including genetic iron overload, suggest that there are yet largely undetermined factors which modulate the disease clinical expression. Among potential factors, we hypothesized that interactions of iron metabolism with other metals could play a role. In a first approach we explored whether the modulation of iron metabolism existing between mice of three genetic backgrounds, was, or not, associated with modulation of non-iron metal parameters.
As expected for iron parameters (Champy et al. 2008; Courselaud et al. 2004; Dupic et al. 2002; Leboeuf et al. 1995; Siegers et al. 1977; Wang et al. 2007), Balb/c and DBA/2 mice presented significantly higher iron concentrations and transferrin saturation values in plasma and increased liver iron concentrations compared to C57BL/6 mice. These results confirm the impact of genetic background on systemic iron metabolism parameters in mice. This concept is also supported by data showing, in drosophila, that Fe concentration varies between species (Sadraie and Missirlis 2011).
Looking for the relationships between the levels of hepatic hepcidin 1 mRNA, which encodes the gate keeper of iron parameters on the one hand, and plasma iron, transferrin saturation and tissue iron concentrations on the other hand, no statistically significant relationship was found. Moreover, DBA/2 mice showed significantly lower hepatic hepcidin 1 mRNA levels than Balb/c mice, whereas the previously reported hepcidin 1 mRNA increase in C57BL/6 versus DBA/2 mice was not found (Coppin et al. 2007). Thus, in these wild type healthy mice, hepcidin levels were closed from one strain to another, despite significant differences in systemic iron parameters.
These data suggest that, in normal healthy condition, physiological parameters, that are independent of iron metabolism, might subtly regulate hepcidin levels and intervene continuously, thus modulating the strength of the relationship between iron parameters and hepcidin expression as reported in iron-related disorders.
They may also support our hypothesis that other, not yet identified parameters, play a role in the control of hepcidin expression and/or iron parameters. Thus, non-iron metals, for which associations with iron metabolism and/or diseases have been reported (Loreal et al. 2014), could participate to the modulation of iron parameters. The links between parameters of these metals, iron metabolism and genetic background have not yet been studied in mice. However, comparing concentrations, it has been reported that Zn, Cu and Mn concentrations may vary between Tephritidae (fruit flies) species fed with their regular diet. In addition those parameters can be changed by modification of regimen (Rempoulakis et al. 2014).
We chose to investigate, in mice receiving the same diet, four metals, Cu, Zn, Mn and Co, for which associations with iron have been reported in human health, and two other (Mg and Rb), which exert potential biological functions and for which interactions with iron have not yet been evaluated. Thus, we found significant variations of metal concentrations which are likely only related to genetic background differences. In addition, when looking at the metal parameter levels in the four matrices of the three strains, the PCA analysis led us to stratify the strains. The largest difference was found between the C57BL/6 and the DBA/2 strains. It is noteworthy that Fe was not the only metal participating to such stratification. On the one hand, one part of the trace elements in some matrices showed similar concentrations between the three mice strains. This observation suggests that these trace elements may obey a general and identical law for each mice strain. On the other hand, part of trace elements in some matrices showed significant differences between mice strains.
Links between plasma concentrations of the different metals are difficult to interpret due to the fact that plasma metal concentrations are temporary conditions which vary due to digestive absorption, but also to different half-lives in the circulation, related to different modalities in tissue uptake and/or elimination. However, it is noteworthy that the only statistical link involving plasma transferrin saturation that we identified concerned plasma copper. Such a relationship was not surprising, knowing that the main copper-linked protein is ceruloplasmin, a multicopper oxidase that plays a major role, through its ferroxidase activity, in the interaction between ferric Fe and transferrin (Mukhopadhyay et al. 1998).
As to the erythrocytes, statistical relationships were found between all the studied metal parameters and are therefore difficult to interpret. A slight statistical link was observed between haemoglobin and hepatic hepcidin 1 mRNA level expression, suggesting that in normal condition tissue oxygenation level and/or erythropoietic activity could be critical factors in the balance of hepcidin expression. The recently reported erythroferrone factor (Kautz et al. 2014) could be involved.
In the liver and spleen, correlations were found between the concentrations of some metals and iron in the corresponding tissue and/or hepatic hepcidin mRNA levels.
Rb, a metal with antidepressant properties (Tuoni et al. 1987), strongly separates C57BL/6 and DBA/2 strains in PCA. Such differences in Rb parameters between strains have been previously suggested (Siegers et al. 1977). Moreover, the linear regression statistical analysis taking the strain into account supports the link between liver or spleen Rb concentrations and hepcidin1 mRNA levels. Quickly absorbed after oral ingestion, Rb is distributed throughout the body and eliminated in urine. The highest concentrations are found in liver and muscles (Meltzer 1991; Tuoni et al. 1987). Rb is taken up by cells through membrane channels shared with potassium, and this property is used to perform cardiac function evaluation by using radioactive 82Rb and positron emission tomography (Mc Ardle et al. 2012). The biologic half-life of Rb is close to 45 days. Chronic exposure to Rb may favour its body accumulation (Bentley and Miller 1977; Meltzer 1991). The link between Rb and hepcidin mRNA level warrants further investigation.
Links were also found between Co and Fe concentrations in liver and spleen. Co exposure in humans results, mainly in developed countries, from prosthetic material in addition to oral ingestion as pollutant. It has been reported that DMT1 is able to take up Co as other divalent cations (Gunshin et al. 1997). Co, by stabilizing the transcriptional factor HIF (hypoxia-inducible factor), may mimic hypoxia and, in turn, stimulates adaptive response to hypoxia (Simonsen et al. 2012). Interactions between iron and Co have been described. It has been initially reported that digestive absorption of iron and Co was increased in rats with iron deficiency (Pollack et al. 1965), in patients with hepatic steatosis complicated by iron deficiency and in subjects with iron deficiency, compared to a similar group with normal iron status (Olatunbosun et al. 1970). Digestive absorption of Co was also increased in patients with genetic hemochromatosis or liver cirrhosis complicated by iron overload (Olatunbosun et al. 1970). Therefore, Co interacts with iron metabolism.
Relationships between Mn and Fe concentrations both in liver and spleen were also found. Mn is a metal which is implicated in enzymatic activities, especially those associated with Mn-SOD which are involved in defense mechanisms against reactive oxygen species (Macmillan-Crow and Cruthirds 2001). Mn is mainly absorbed from digestive lumen but may also be acquired by airway. The first hepatic passage is strongly involved in Mn elimination (Cotzias 1958; Papavasiliou et al. 1966). Hepatic dysfunction has been reported favoring encephalopathy related to Mn excess in brain (Butterworth 2013). As for cobalt, it has been reported that digestive absorption of Mn and iron was increased in rats with iron deficiency (Pollack et al. 1965). It is noteworthy that in Hfe−/− mice, a model of hemochromatosis, Mn excess was associated with the iron overload, thus strongly linking the two metals in an iron-related disorder (Kim et al. 2013). Moreover, the data obtained in healthy animals indicates that these two metals are also associated in normal situations.
In conclusion, the present data shows that genetic background strongly modulates metal parameters. Moreover, they indicate that links could exist between iron parameters and non-iron metals parameters. This suggests that modulation of non-iron metals may interfere with iron metabolism in health and diseases. Therefore, these data indicate that knowledge on iron metabolism will benefit from further studies on the relationships between iron and other metals, especially when considering the increasing exposure of individuals to various non-iron metals during occupational or personal life.
References
An P et al (2012) TMPRSS6, but not TF, TFR2 or BMP2 variants are associated with increased risk of iron-deficiency anemia. Hum Mol Genet 21:2124–2131. doi:10.1093/hmg/dds028
Andrews NC (1999) Disorders of iron metabolism. N Engl J Med (published erratum appears in N Engl J Med 2000 Feb 3;342(5):364) 341:1986–1995
Balesaria S, Ramesh B, McArdle H, Bayele HK, Srai SK (2010) Divalent metal-dependent regulation of hepcidin expression by MTF-1. FEBS Lett 584:719–725. doi:10.1016/j.febslet.2009.12.023
Bentley SA, Miller DT (1977) Radionucleide blood survival studies. In: Lewis SM (ed) Radioisotopes in haematology. Sauders, London, pp 245–262
Brissot P, Troadec MB, Bardou-Jacquet E, Le Lan C, Jouanolle AM, Deugnier Y, Loreal O (2008) Current approach to hemochromatosis. Blood Rev 22:195–210. doi:10.1016/j.blre.2008.03.001
Butterworth RF (2013) The liver-brain axis in liver failure: neuroinflammation and encephalopathy. Nat Rev Gastroenterol Hepatol 10:522–528. doi:10.1038/nrgastro.2013.99
Camaschella C et al (2000) The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet 25:14–15
Carroll BJ, Sharp PT (1971) Rubidium and lithium: opposite effects on amine-mediated excitement. Science 172:1355–1357
Champy MF et al (2008) Genetic background determines metabolic phenotypes in the mouse. Mamm Genome 19:318–331. doi:10.1007/s00335-008-9107-z
Coppin H et al (2007) Gene expression profiling of Hfe−/− liver and duodenum in mouse strains with differing susceptibilities to iron loading: identification of transcriptional regulatory targets of Hfe and potential hemochromatosis modifiers. Genome Biol 8:R221. doi:10.1186/gb-2007-8-10-r221
Cotzias GC (1958) Manganese in health and disease. Physiol Rev 38:503–532
Courselaud B et al (2004) Strain and gender modulate hepatic hepcidin 1 and 2 mRNA expression in mice. Blood Cells Mol Dis 32:283–289. doi:10.1016/j.bcmd.2003.11.003
Dupic F et al (2002) Inactivation of the hemochromatosis gene differentially regulates duodenal expression of iron-related mRNAs between mouse strains. Gastroenterology 122:745–751
Fawcett WJ, Haxby EJ, Male DA (1999) Magnesium: physiology and pharmacology. Br J Anaesth 83:302–320
Finberg KE et al (2008) Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat Genet 40:569–571
Finberg KE, Whittlesey RL, Andrews NC (2011) Tmprss6 is a genetic modifier of the Hfe-hemochromatosis phenotype in mice. Blood 117:4590–4599. doi:10.1182/blood-2010-10-315507
Garcia SJ, Gellein K, Syversen T, Aschner M (2007) Iron deficient and manganese supplemented diets alter metals and transporters in the developing rat brain. Toxicol Sci 95:205–214. doi:10.1093/toxsci/kfl139
Gunshin H et al (1997) Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 388:482–488
Hansen SL, Trakooljul N, Liu HC, Hicks JA, Ashwell MS, Spears JW (2010) Proteins involved in iron metabolism in beef cattle are affected by copper deficiency in combination with high dietary manganese, but not by copper deficiency alone. J Anim Sci 88:275–283. doi:10.2527/jas.2009-1846
Harris ZL, Takahashi Y, Miyajima H, Serizawa M, MacGillivray RT, Gitlin JD (1995) Aceruloplasminemia: molecular characterization of this disorder of iron metabolism. Proc Natl Acad Sci USA 92:2539–2543
Island ML, Jouanolle AM, Mosser A, Deugnier Y, David V, Brissot P, Loreal O (2009) A new mutation in the hepcidin promoter impairs its BMP response and contributes to a severe phenotype in HFE related hemochromatosis. Haematologica 94:720–724. doi:10.3324/haematol.2008.001784
Kautz L, Jung G, Valore EV, Rivella S, Nemeth E, Ganz T (2014) Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat Genet 46:678–684. doi:10.1038/ng.2996
Kim J, Buckett PD, Wessling-Resnick M (2013) Absorption of manganese and iron in a mouse model of hemochromatosis. PLoS ONE 8:e64944. doi:10.1371/journal.pone.0064944
Latour C et al (2014) Testosterone perturbs systemic iron balance through activation of epidermal growth factor receptor signaling in the liver and repression of hepcidin. Hepatology 59:683–694. doi:10.1002/hep.26648
Leboeuf RC, Tolson D, Heinecke JW (1995) Dissociation between tissue iron concentrations and transferrin saturation among inbred mouse strains. J Lab Clin Med 126(2):128–136
Le Gac G et al (2004) The recently identified type 2A juvenile haemochromatosis gene (HJV), a second candidate modifier of the C282Y homozygous phenotype. Hum Mol Genet 13:1913–1918
Loreal O et al (2002) Aceruloplasminemia: new clinical, pathophysiological and therapeutic insights. J Hepatol 36:851–856
Loreal O, Cavey T, Bardou-Jacquet E, Guggenbuhl P, Ropert M, Brissot P (2014) Iron, hepcidin, and the metal connection. Front Pharmacol 5:128. doi:10.3389/fphar.2014.00128
Macmillan-Crow LA, Cruthirds DL (2001) Invited review: manganese superoxide dismutase in disease. Free Radic Res 34:325–336
Mc Ardle BA, Dowsley TF, deKemp RA, Wells GA, Beanlands RS (2012) Does rubidium-82 PET have superior accuracy to SPECT perfusion imaging for the diagnosis of obstructive coronary disease?: A systematic review and meta-analysis. J Am Coll Cardiol 60:1828–1837. doi:10.1016/j.jacc.2012.07.038
Melis MA et al (2008) A mutation in the TMPRSS6 gene, encoding a transmembrane serine protease that suppresses hepcidin production, in familial iron deficiency anemia refractory to oral iron. Haematologica 93:1473–1479
Meltzer HL (1991) A pharmacokinetic analysis of long-term administration of rubidium chloride. J Clin Pharmacol 31:179–184
Merryweather-Clarke AT et al (2003) Digenic inheritance of mutations in HAMP and HFE results in different types of haemochromatosis. Hum Mol Genet 12:2241–2247
Milet J et al (2007) Common variants in the BMP2, BMP4, and HJV genes of the hepcidin regulation pathway modulate HFE hemochromatosis penetrance. Am J Hum Genet 81:799–807. doi:10.1086/520001
Mitchell CJ, Shawki A, Ganz T, Nemeth E, Mackenzie B (2014) Functional properties of human ferroportin, a cellular iron exporter reactive also with cobalt and zinc. Am J Physiol Cell Physiol 306:C450–C459. doi:10.1152/ajpcell.00348.2013
Mukhopadhyay CK, Attieh ZK, Fox PL (1998) Role of ceruloplasmin in cellular iron uptake. Science 279:714–717
Nai A et al (2011) TMPRSS6 rs855791 modulates hepcidin transcription in vitro and serum hepcidin levels in normal individuals. Blood 118:4459–4462. doi:10.1182/blood-2011-06-364034
Olatunbosun D, Corbett WE, Ludwig J, Valberg LS (1970) Alteration of cobalt absorption in portal cirrhosis and idiopathic hemochromatosis. J Lab Clin Med 75:754–762
Papanikolaou G et al (2004) Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet 36:77–82. doi:10.1038/ng1274ng1274
Papavasiliou PS, Miller ST, Cotzias GC (1966) Role of liver in regulating distribution and excretion of manganese. Am J Physiol 211:211–216
Pollack S, George JN, Reba RC, Kaufman RM, Crosby WH (1965) The absorption of nonferrous metals in iron deficiency. J Clin Invest 44:1470–1473. doi:10.1172/JCI105253
Rempoulakis P et al (2014) Conserved metallomics in two insect families evolving separately for a hundred million years. Biometals 27:1323–1335. doi:10.1007/s10534-014-9793-9
Roetto A et al (2003) Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet 33:21–22
Sadraie M, Missirlis F (2011) Evidence for evolutionary constraints in Drosophila metal biology. Biometals 24:679–686. doi:10.1007/s10534-011-9420-y
Siegers MP, Kasperek K, Heiniger HJ, Feinendegen LE (1977) Distribution of trace elements in organs of mice of different inbred strains. J Radioanal Chem 37:421–426
Simonsen LO, Harbak H, Bennekou P (2012) Cobalt metabolism and toxicology: a brief update. Sci Total Environ 432:210–215. doi:10.1016/j.scitotenv.2012.06.009
Troadec MB, Ward DM, Lo E, Kaplan J, De Domenico I (2010) Induction of FPN1 transcription by MTF-1 reveals a role for ferroportin in transition metal efflux. Blood 116:4657–4664. doi:10.1182/blood-2010-04-278614
Tuoni M et al (1987) Renal tolerance of rubidium chloride: short-term clinical evaluation. J Clin Pharmacol 27:503–507
Videt-Gibou D et al (2009) Iron excess treatable by copper supplementation in acquired aceruloplasminemia: a new form of secondary human iron overload? Blood 114:2360–2361. doi:10.1182/blood-2009-06-226175
Wang F et al (2007) Genetic variation in Mon1a affects protein trafficking and modifies macrophage iron loading in mice. Nat Genet 39(8):1025–1032
Weiss G, Goodnough LT (2005) Anemia of chronic disease. N Engl J Med 352:1011–1023
Yamaguchi S et al (2007) Effects of lead, molybdenum, rubidium, arsenic and organochlorines on spermatogenesis in fish: monitoring at Mekong Delta area and in vitro experiment. Aquat Toxicol 83:43–51. doi:10.1016/j.aquatox.2007.03.010
Acknowledgments
This work was supported by INSERM and University of Rennes 1.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cavey, T., Ropert, M., de Tayrac, M. et al. Mouse genetic background impacts both on iron and non-iron metals parameters and on their relationships. Biometals 28, 733–743 (2015). https://doi.org/10.1007/s10534-015-9862-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-015-9862-8