Abstract
As an effective alternative for dissolved nitrogen removal, biofilter closely associates its treatment performance to structural and/or operational conditions. In this study, a set of four different biofilters including MAVF (mature aerated vertical flow), NAVF (new aerated vertical flow), NVF (new non-aerated vertical flow), and BHF (baffled non-aerated horizontal flow) were employed to purify low C/N ratio (3.8) domestic wastewater. All the filters were packed with round ceramsite operated under varying hydraulic loading rates (HLRs) of 0.024–0.18 m/day. During the start-up, both the physicochemical and microbial characterizations were investigated. It was found that, carbon and nitrogen could achieve ideal removal in MAVF once added with further sedimentation, while phosphorus displayed an unsatisfactory sequestration in any of the four filters probably due to the high inflow load and/or lack of alternate anaerobic/aerobic conditions. Filter clustering based on percent removal and removal rate constant displayed a consistent pattern, which was similar to that based on taxa of phylum from 16S rRNA sequencing, or phylum/genus/species from shotgun metagenomic sequencing although there were obvious distinctions in taxa compositions among direct comparison. Meanwhile, gene function annotation revealed that filter clustering based on metabolic pathways was consistent with that based on purification performance. These consistencies might imply that the treatment performance was mainly determined by microbial degradation. The enrichment of specific functional microbes responsible for the degradation of certain pollutants, such as carbohydrates, matched well with the defined purification performance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nowadays under the increasing pressure of environmental protection, many of China’s wastewater treatment plants (WWTPs) face to supplement advanced treatments to meet the first A standards (GB 18,918–2002, China) (Qu et al. 2019). Among the advanced treatments, dissolved nitrogen removal is still the key that can mostly be achieved in biological ways. As a mature biofilm technology, biofiltration has been widely applied for dissolved nitrogen removal (Garcia-Ruiz et al. 2018; Jiang et al. 2018; Lin et al. 2019). The main processes involve nitrification and denitrification completed by nitrifiers and denitrifiers, respectively. These functional microbes are highly susceptible to many factors, such as carbon sources (He et al. 2018b), dissolved oxygen (DO) (Jiang et al. 2018; Rout et al. 2017), and pH (Jiang et al. 2018). To achieve an ideal performance, it is essential to unravel the complicated biological processes from the perspective of microbial characterizations.
Currently, metagenomics is a powerful approach in deciphering microbial characterizations. It can not only present compositional/phylogenetic characterizations but also display functional profilings (Chu et al. 2018; Ng et al. 2019; Tang et al. 2016). According to different purposes, metagenomics can achieve both qualitative and quantitative analyses of specific microorganisms via targeted or untargeted ways. In targeted ways, such as the cost-effective 16S rRNA sequencing, a gene or a few genes are sequenced, while in untargeted ways, such as the high-cost shotgun metagenomic sequencing, all present DNA is sequenced (Garrido-Cardenas et al. 2017). At present, the 16S rRNA sequencing is frequently used for structural comparison (Garrido-Cardenas et al. 2017), while the shotgun metagenomic sequencing is widely applied for functional clarification (Jadeja et al. 2014).
Recently many studies have adopted these two approaches to decipher microbial characterizations (Bai et al. 2017; Guan et al. 2018; Tang et al. 2016; Zhao et al. 2019). However, due to different data processing, both consistent and conflict results were reported. For the examples of consistency, RiboFR-Seq linked the annotations of 16S rRNA and metagenomic contigs to make a consistent classification (Zhang et al. 2016b); Tax4Fun provided a good approximation to functional profiles (Asshauer et al. 2015); 16S rRNA gene sequences provided results comparable to shotgun metagenomic sequencing (Mitra et al. 2013). However, for the instance of conflict, 16S rRNA and shotgun metagenomic sequencing revealed a distinct microbial community profile (Delforno et al. 2017). The above cases indicate that the consistency mainly focuses on the functional classifications, while the conflict mainly focuses on the direct comparison of microbial community. However, up to date, there is still little information on the comparison of statistical associations of biofilter performance with microbial characterizations, which are unraveled by both 16S rRNA and shotgun metagenomic sequencing.
So in this study, both of the above two approaches were employed to unravel the microbial characterizations of four different biofilters, which were used to treat simulated domestic wastewater under different conditions referring aeration, microbial acclimation period, pool shape, and inflow loading. The purposes of the study were to verify: (1) Whether there was an obvious distinction in microbial characterizations between 16S rRNA and shotgun metagenomic sequencing? (2) Did the microbial structural characterizations revealed by the two methods consistently associate with treatment performance? (3) In the same way, did the microbial functional characterizations unraveled by shotgun metagenomic sequencing consistently associate with treatment performance? Through the above analyses, a better understanding of the complicated links of microbial characterizations to purification performance would be obtained, which might be helpful for biofilter optimization or management.
Materials and methods
System construction
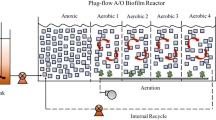
For the present study, four plexiglass biofilters including three vertical flow filters (named as MAVF, NAVF & NVF) and one baffled horizontal flow (BHF) were constructed. The baffled flow was achieved by installing nine parallel baffles (39 cm L × 0.9 cm W × 48 cm H for each one). The scale of the vertical flow filter was 48 cm in length (L) × 48 cm in width (W) × 60 cm in height (H) and that of the horizontal flow was 100 cm L × 48 cm W × 48 cm H. All the filters were packed with round ceramsite with a diameter of 3–5 mm, a porosity of 0.433, and a depth of 38 cm and 34 cm for the vertical and horizontal flow filters, respectively. To ward off influent pump flushing, perforated water distribution pipes (made of PVC, Φ16 mm) were installed on the top of the vertical flow filter; meanwhile, nanoporous aeration tubes (rubber, Φ15 mm) were fixed in perforated casing pipes (PVC, Φ20mm) at the bottom. The nanoporous tubes were then linked to an air compressor (power: 250 W; blowing rate: 60 m3/h; maximum pressure: 10 kPa; model: HG-250) forming an aeration system. Finally, an outlet tap was set up at the bottom of each vertical flow filter and at the upper edge of the horizontal flow one to regulate water drainage.
Experimental design
In this study, all the filters were used to treat simulated domestic wastewater, whose constituents are listed in Table 1. The preparation scheme was as follows: KH2PO4 0.0439 g/L; NH4HCO3 0.2256 g/L; C6H12O6 0.15 g/L; wheat flour 0.3 g/L. The scheme was referred to an effluent from the grit chamber of a local WWTP. The wastewater was characterized by a low C/N ratio (3.8) and was dominated by dissolved ammonium and phosphorus. To expand the difference among the four filters, the MAVF and BHF constituting a combination system had been used to treat simulated high-strength aquaculture wastewater for over a year (Zhang et al. 2017) therefore possessing mature biofilm, while the NAVF (representing new aerated vertical flow filter) and NVF (new non-aerated vertical flow filter) were new ones.
Usually, for a mature biofilter, its performance is strongly associated with operating conditions. However, for a new biofilter, its performance may be determined by to what extent the biofilm has developed. No matter either operating conditions or biofilm maturation, they mainly affect biofilter performance via shaping different microbial communities. Up to date, the reports of suitable start-up time for new biofilters mostly ranged from few days to more than a month, such as the volcanic carrier biofilter controlled based on online pH-DO linkage control strategy (Jiang et al. 2018), the lab-scale biofilters inoculated with nitrifying and backwashing sludge (Cai et al. 2015), and the bench-scale Filtralite biofilters operated under anoxic conditions (Garcia-Ruiz et al. 2018). In this study, due to the medium wastewater concentration (Table 1), the suitable start-up time was assumed a month, during which three-stage batch tests were performed. That is the HLRs of the three vertical flow filters were adjusted to be 0.05, 0.10, and 0.18 m/day in three stages with each one lasting 10 days. Accordingly, the HLRs of the BHF were 0.024, 0.048, and 0.086 m/day, respectively.
For a rapid start-up, the four filters were daily inoculated with activated sludge taken from the above-mentioned WWTP for 3 days. The physicochemical characteristics of the inoculated activated sludge are listed in Table 2, which showed that dissolved nutrients were mainly incorporated into microbial biomass that was beneficial for inoculation. Afterward, the MAVF, NAVF, and NVF were fed with the simulated domestic wastewater intermittently once a day, while the BHF only received outflow of the MAVF. Before each feeding, the three vertical flow filters were completely drained and then fed again to the designated water level (corresponding to each HLR). Meanwhile, the MAVF and NAVF performed intermittent aeration while the BHF and NVF did not. The aeration covered a total of 6 h per day including two sections, i.e., 09:00–12:00 a.m. and 14:30–17:30 p.m. Accordingly, the gas/water ratios corresponding to the above three HLRs were about 1200:1, 600:1, and 300:1, respectively. Hereby, it could be considered that, the four filters had been characterized by different structural and/or operational conditions that would produce a distinction in treatment performance, which might be mainly caused by different microbial communities.
Physicochemical analysis
Water samples for physicochemical analysis were collected daily and the sampling sites included inflow and outflow. The inflow was sampled instantly once the simulated domestic wastewater had been prepared in a feeding tank, while the outflow was sampled on the next day during the draining process. The monitored parameters included online and laboratory indices. The online indices included temperature (T), dissolved oxygen (DO), redox potential (ORP), alkalinity (pH), electronic conductivity (EC), total soluble solids (TDS) and salinity, which were measured in situ by a multi-parameter water quality analyzer (Model: YSI Pro Plus), while the laboratory parameters involved chemical oxygen demand (COD), total ammonium nitrogen (TAN), nitrate (NO3−–N), nitrite (NO2−–N), total nitrogen (TN), total phosphorus (TP), and inorganic phosphorus (IP), which were measured according to the standard methods (APHA 2005). The COD also included total content (TCOD) and the dissolved portion (DCOD). The TCOD was determined directly while the DCOD was measured after the water sample had been filtered through a 0.45 μm fiber membrane.
Microbial analysis
At the end of batch tests, the ceramsite inside each filter was sampled, conserved and sent to LC-Bio Technology Co., Ltd (Hangzhou, China) for microbial analysis, which covered both 16S rRNA and shotgun metagenomic sequencing. DNA was extracted respectively from each sample using the E.Z.N.A.® Stool DNA Kit (D4015-02, Omega, Inc., USA) according to the manufacturer’s instructions. The reagent had been proven effective when designed to uncover DNA from trace amounts of sample. The total DNA was eluted in 50 µL of Elution buffer and stored at − 80 °C until measurement. For 16S rRNA sequencing, the V3–V4 region of the prokaryotic small-subunit (16S) rRNA gene was amplified with slightly modified versions of primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). PCR amplification was performed following the methods given by Wang et al. (2019) with final products confirmed with 2% agarose gel electrophoresis, purified by AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, USA), and quantified by Qubit ( Invitrogen, USA). After then, samples were sequenced on an Illumina MiSeq platform according to the manufacturer's recommendations. Subsequent data analysis was done as the methods described by Tao et al. (2019). Similarly, for shotgun metagenomic sequencing, DNA library construction and subsequent data analysis were completed according to the methods depicted by He et al. (2018a). For the present study, gene function was annotated according to databases of GO (gene ontology), KEGG (Kyoto encyclopedia of genes and genomes), eggNOG (evolutionary genealogy of genes: non-supervised orthologous groups) and CAZy (carbohydrate-active enzymes). GO is a widely used bioinformatics concept that unifies genes and their products of all species. KEGG can identify metabolic pathways. EggNOG can classify the potential functions of unigenes. CAZy can classify the functions of carbohydrate-active enzymes (Cai et al. 2018).
Statistical analysis
In this study, data were expressed as mean ± standard deviation (SD). Total inorganic nitrogen (TIN) was defined as the sum of TAN + NO3−–N + NO2−–N, while organic nitrogen (Org-N) was defined as the deduction of TN-TIN. Treatment performance was evaluated by percent removal and first-order removal rate constant K with the latter calculated by the following formula (Zhang et al. 2016a):
where K was the first-order removal rate constant, day−1; HLR was the hydraulic loading rate, m/day; Cin and Cout were the inflow and outflow concentrations, respectively, mg/L; hw was the filling depth, m; ε was the porosity.
An independent t-test was used to detect the differences in physicochemical parameters between inflow and outflow as well as between aeration and non-aeration. Similarly, covariance analysis was selected to compare the differences in percent removal/removal rate constant K among different filters with inflow concentration as a covariate. This was because the inflow of BHF was the outflow of MAVF. Meanwhile, multiple comparisons were performed with LSD for equal variances and Tamhane's T2 for unequal variances. Significant differences (P < 0.05) among the four filters were marked with different letters.
Results and discussion
Comparison of online physicochemical parameters
Table 3 shows that the significant differences in physicochemical parameters between inflow and outflow of the four different biofilters mainly focused on temperature, DO, ORP, and pH, while only significant difference was observed in MAVF for SC, TDS and salinity. Besides, the mechanical aeration significantly increased outflow DO and pH (i.e., by comparing NVF with NAVF/MAVF with all P < 0.05). The reason for the increase of DO was due to that, the nitrification process, as well as the oxidation of organics, consumed DO that could be supplemented via mechanical aeration in aerated filters. However, in non-aerated filter, the consumed DO was mainly derived from feeding wastewater apart from the limited atmospheric diffusion. Similarly, the reason for the increase of pH was possibly due to that, denitrification process produced CO2, which might be partially blown out by the intermittent aeration in aerated filters, while in non-aerated filter, it might retain in the system leading to a lower pH value (Zhang et al. 2019b). Finally, according to the thresholds for DO classification proposed by Gao et al. (2007), namely < 0.3 mg/L representing for anaerobic, 0.4–0.7 mg/L for facultative, and > 1.0 mg/L for aerobic, the DO content in the four filters was at facultative/aerobic level meaning the lack of alternate anaerobic/aerobic conditions in a single reactor.
Comparison of treatment performance
According to Table 4, mean outflow concentrations of NO3−–N and NO2−–N in each filter were no more than 0.40 mg/L and 0.02 mg/L, respectively, indicating no obvious accumulation of nitrate or nitrite. This also demonstrated that complete denitrification occurred inside the four filters. Accordingly, DIN in the outflow of each filter was mainly composed of unnitrified TAN. Besides, TN in the outflow of MAVF/BHF was mainly composed of Org-N while DIN accounted for a major portion in TN in the outflow of NAVF/NVF. This might be explained by the fact that, the MAVF/BHF were more mature than the NAVF/NVF thus possessing higher biofilm biomass. The relatively high content of Org-N in the outflow of MAVF/BHF was mainly composed of fallen biofilm. Thus, TN removal in mature filter could be further improved by sedimentation to reach the first A standard (< 15 mg/L, GB 18,918–2002, China). Although the NAVF was aerated, its nitrifying capacity was much lower compared to the MAVF indicating biofilm maturation contributed a lot to nitrification.
From the perspective of percent removal for each parameter, the MAVF, NAVF, and NVF all displayed high removal efficiency on organics (mean TCOD percent removal > 80%), while the BHF only displayed positive removal on organics and Org-N (Table 4). The positive removal of organics, Org-N and TN as well as the negative removal of TAN, NO3–−N, NO2−–N and DIN in the BHF indicated that, particle sequestration, ammonification and ammonium/nitrite oxidation occurred simultaneously inside the filter bed. Meanwhile, after compared the concentration of each nitrogen in inflow/outflow, it was deemed that the above nitrogen conversion intensity in BHF was extremely weak, which might be mainly attributed to the relatively low DO inside the filter bed (Jiang et al. 2018; Zhang et al. 2019b). However, for phosphorus removal, the four filters all displayed low removal efficiency resulting in high outflow concentrations. This was probably due to the high inflow load (mean inflow TP = 9.54 mg/L, Table 1) and the lack of alternate anaerobic/aerobic conditions inside the filter bed (He et al. 2018b).
Similarly, from the perspective of removal rate constant K for each parameter, the MAVF, NAVF, and NVF all presented relatively high K for nitrate (Table 4) implying complete denitrification occurred inside these systems. It was probably because denitrification was easy to happen once guaranteed with sufficient endogenous carbon sources derived from the fallen biofilm and/or inflow load (TCOD/TN = 3.8, Table 1), and the denitrification had not been inhibited by the intermittent aeration. On the contrary, the various K of each nitrogen were almost negative (except TN and Org-N) in BHF implying nitrification and denitrification were both greatly restrained. This was probably attributed to the anoxic (or even anaerobic) environment inside the filter bed (Zhang et al. 2019b) that was adverse to nitrification and thus could not supply additional nitrate/nitrite for denitrification. Besides, the inflow of BHF (namely MAVF outflow) contained an extremely low level of carbon source (mean DCOD = 12 mg/L, Table 4) and nitrate/nitrite (mean NO2−–N = 0.01 mg/L, mean NO3−–N = 0.1 mg/L) that was also detrimental to denitrification (He et al. 2018b; Zhang et al. 2018). Covariance analysis showed that there were significant differences in K of each parameter among the four filters. The overall rank of purification efficiency among the four filters was MAVF > NAVF > NVF > BHF. Finally, filter clustering based on percent removal and removal rate constant K showed a consistent pattern over the four filters. That is the NAVF and NVF showed the closest distance followed by MAVF, while the BHF was the farthest from the others (Fig. 1).
Heatmap comparison of treatment performance a based on percent removal and b based on removal rate constant K among the four different biofilters. The blue and red colors denote low and high values, respectively. Euclidean distance is computed to evaluate the similarity between filters, and clustering is conducted using complete linkage method. The same for the following. (Color figure online)
Comparison of microbial composition
After database alignment through characteristic sequences, the taxa richness at the phylum/genus/species level annotated from shotgun metagenomic sequencing was extremely higher than that annotated from 16S rRNA sequencing (Table 5). Unlike the 16S rRNA sequencing, the shotgun metagenomic sequencing could annotate most of the dominant taxa to specific names at the species level. Because listing all taxa names to analyze microbial community was unrealistic due to their huge richness (Table 5), and dominant taxa could generally reflect the main characterizations of microbial community (Wang et al. 2018). Therefore, the top 20 taxa with the highest abundance were picked out, and the rest was merged to others to express relative abundance, which was used for subsequent heatmap analysis. For the present study, the heatmaps were constructed at three levels, i.e., phylum, genus, and species (Fig. 2), because most of them were frequently depicted in similar studies for microbial comparison (Du et al. 2017; Gu et al. 2019; He et al. 2018b).
According to 16S rRNA sequencing, there were obvious distinctions in dominant taxa compositions at any level of phylum/genus/species. Taking the phylum for example, the NAVF and NVF, compared to MAVF and BHF, had a relatively higher abundance of Verrucomicrobia and Actinobacteria, while got a lower abundance of Acidobacteria, Chlorobi and Candidatus Saccharibacteria, etc. These distinctions resulted in a clustering of NAVF and NVF being the closest, while BHF being the farthest from the remaining filters (Fig. 2a). The clustering pattern was consistent with purification performance as shown in Fig. 1. Nevertheless, at the genus or species level, a different clustering pattern was observed compared to treatment performance (Fig. 2c, e). Besides, most of these taxa had not been identified to species level and just been named at the genus level. Since most of the dominant taxa shared the common name at both genus and species levels, filter clustering displayed a similar pattern at the two levels.
Similarly, according to shotgun metagenomic sequencing, there were also obvious distinctions in dominant taxa compositions at any level of phylum/genus/species over the four filters. Taking the phylum for example, Firmicutes was the most abundant taxa in BHF, followed by NVF, MAVF, and NAVF in a descending order (Fig. 2b). Nevertheless, filter clustering based on the three levels of phylum, genus, and species all displayed a similar pattern (Fig. 2b, d, f), which was consistent with treatment performance as shown in Fig. 1. This demonstrated that the dominant taxa annotated from shotgun metagenomic sequencing were more closely related to treatment performance compared to those annotated from 16S rRNA sequencing. The reason was probably due to that most of the dominant taxa annotated from shotgun metagenomic sequencing had the function of nutrient removal. Taking the genus for example, some of the genera, such as Bacillus (Du et al. 2018), Pseudomonas (Wang et al. 2018), Brevundimonas (Du et al. 2017), Flavobacterium (Chen et al. 2018), Dechloromonas (Lin et al. 2019), Arthrobacter (He et al. 2017), and Bradyrhizobium (Zou et al. 2018), had been frequently reported as facultative nitrifiers and/or denitrifiers.
Comparison of gene function annotated by different databases
Gene function annotated by GO
GO is a database established by the Federation of Gene Ontology, aiming at establishing a standard applicable to all species. GO provides three kinds of systematic definitions for describing the functions of gene products, i.e., GO function, GO ID, and GO term. In the present study, the first two definitions were analyzed.
According to GO function, the molecular function genes showed the highest abundance, followed by the biological process genes. The cellular component genes showed the lowest abundance. Besides, a considerable quantity of genes was not annotated (i.e., NA in Fig. 3a, the same for the following). Similarly, according to GO ID, the total of other low abundant genes was the highest, followed by the non-annotated genes. GO:0,005,575, GO:0,003,674, and GO:0,008,150 showed a higher abundance than the rest (Fig. 3b). Filter clustering based on GO ID showed a different pattern compared to that based on GO function. Nevertheless, both clustering patterns were different from that based on purification performance (Fig. 1).
Gene function annotated by KEGG
KEGG is a database that links genome molecular level information with high-level biological system function information. KEGG can be divided into 17 main databases, among which the KEGG pathway is widely used for annotation of genome and high-throughput data. In the present study, the KEGG pathway was adopted to describe the functions of gene products from four systematic definitions, i.e., KEGG level 1, level 2, pathway entry, and KO entry.
According to KEGG pathway classification, the most abundant genes were belonged to metabolism, followed by environmental information processing, and genetic information processing. Genes belonged to organismal systems and human diseases showed the least abundance. Among the metabolism, the most abundant genes were belonged to carbohydrate metabolism, followed by overview, amino acid metabolism, metabolism of cofactors and vitamins, and energy metabolism (Fig. 4a). This was because the organics in the raw wastewater were only composed of wheat flour and glucose, both of which are belonged to carbohydrates. The degradation of these carbohydrates was also related to amino acid and energy metabolism via microbial processing (Cerrone et al. 2011). This had also been demonstrated by the coexisting genes of glycan biosynthesis and metabolism (Fig. 4a). Besides, a considerable quantity of genes, which were belonged to biosynthesis of other secondary metabolites, as well as to xenobiotics biodegradation and metabolism, were annotated suggesting that pollutant degradation produced secondary soluble microbial products, which could be used for potential denitrification as endogenous carbon sources (He et al. 2018b; Lin et al. 2019).
Among the environmental information processing, genes belonged to signal transduction showed the highest abundance, followed by membrane transport. This might be due to the activity of glycan biosynthesis and metabolism (Miao et al. 2019) when using glucose as part of the original carbon sources (Miao et al. 2016). Among the cellular processes, cell motility showed the highest abundance. This was probably attributed to the considerable portion of mobile Chloroflexi detected in the reactors (Fig. 2a). Chloroflexi had been frequently reported as a dominant taxa in many similar studies, such as in activated sludge of municipal wastewater treatment plant (Zhang et al. 2019a), in expanded granular sludge bed (EGSB) treating starch-containing wastewater (Qin et al. 2019), and in deep municipal tail wastewater treatment system (Chen et al. 2019). Finally, among the human diseases, genes belonged to drug resistance showed the most abundant. This was probably due to that, many of the taxa, such as Massilia (Wang et al. 2016), Acinetobacter (Yang et al. 2015), Sphingobacterium (Jin et al. 2013), Sphingobium (Zhao et al. 2017), and Leclercia (Sarma et al. 2010), also had the function of aromatics degradation since numerous drugs themselves were belonged to aromatics. These aromatic-degrading microorganisms might be introduced into the reactor with the original sludge inoculation.
Filter clustering based on the four different definitions of the KEGG pathway displayed two distinct patterns: i.e., KEGG level 1 and level 2 displayed a similar pattern (Fig. 4b, c), while the remaining two definitions shared another (Fig. 4d, e). The former pattern was the same as that shown in Fig. 3b, while the latter pattern was the same as purification performance shown in Fig. 1. According to the above descriptions, KEGG level 1 and level 2 denote classification of functional categories, while pathway entry and KO entry denote specific metabolic pathways based on protein ID being obtained in two different manners. Therefore, filter clustering based on gene metabolic pathway was more close to purification performance clustering compared to gene functional classification. This was because the defined purification performance was mainly assessed by pollutant degradation, which was closely related to specific metabolic pathways of certain pollutants, such as carbohydrates (He et al. 2018b).
Gene function annotated by eggNOG
EggNOG is a database of genomic homologous proteins and their functional annotations constructed by the European Molecular Biology Laboratory. EggNOG can be divided into four levels. The first level includes (1) information storage and processing, (2) cellular processes and signaling, (3) metabolism, and (4) poorly characterized. The second level is further subdivided into 25 categories, each of which can be represented by a single letter. The third level is the consensus functional description. The fourth level is specific homologous proteins. For the present study, the latter three levels were used for functional description and clustering.
According to the statistics of all functional gene categories, S denoting function unknown showed the most abundant, followed by E, L, C, P, T, K, M, J, G, etc. (Fig. 5a), in a descending order. The dominant categories of E (representing amino acid transport and metabolism), L (replication, recombination, and repair), C (energy production and conversion), P (inorganic ion transport and metabolism), T (signal transduction mechanisms), J (translation, ribosomal structure, and biogenesis), and G (carbohydrate transport and metabolism) showed similar functions to those annotated by KEGG shown in Fig. 4c. These functions were mainly characterized by substance transport and metabolism accompanied by energy production and conversion. Besides, there were some other categories, such as F (nucleotide transport and metabolism), I (lipid transport and metabolism), N (cell motility), and Q (secondary metabolites biosynthesis, transport, and catabolism), displaying similar functions to those annotated by KEGG (Fig. 4c), too. While the category of A (RNA processing and modification), B (chromatin structure and dynamics), W (extracellular structures), Y (nuclear structure), and Z (cytoskeleton) showed the least abundance that had not been annotated by KEGG. Hereby, it could be considered that the two different annotation methods shared almost similar functions.
Similarly, filter clustering based on functional categories, functional description, and homologous proteins also displayed two distinct patterns: i.e., functional description and homologous proteins shared a similar pattern, which was different from the one clustered by functional categories (Fig. 5b–d). The former pattern was consistent with purification performance clustered shown in Fig. 1, while the latter was similar to that shown in Figs. 3b and 4b, c. Similar to KEGG annotation, the functional description and homologous proteins represented specific metabolic pathways, while the functional categories only denoted their classification. Therefore, it could also be considered that filter clustering was more closely associated with metabolic pathways than with functional classification.
Gene function annotated by CAZy
CAZy is a special database dedicated to the analysis of genome, structure, and biochemical information of carbohydrate-activated enzymes. CAZy database corresponding to level 1 mainly covers six functional categories: glycoside hydrolases (GHs), glycosyltransferases (GTs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), auxiliary activities (AAs), and carbohydrate-binding modules (CBMs) (Yang et al. 2018). The six functional categories can be further divided into functional sub-categories corresponding to level 2.
According to the statistics of all functional genes, GTs showed the most abundant, followed GHs, CBMs, CEs, etc. (Fig. 6a). The AAs and PLs showed the least abundance. The most abundant genes belonged to GTs, GHs and CBMs were probably associated with glycan biosynthesis and metabolism (as shown in Fig. 4c), which was usually accompanied by energy production and conversion (Fig. 5a) (Miao et al. 2016). Before the present study, the four filters had been inoculated with activated sludge taken from a local WWTP for rapid start-up, so they certainly contained glycogen accumulating organisms (GAOs) as demonstrated in Fig. 2. Since the raw wastewater only contained wheat flour and glucose as carbon sources, these carbohydrates could be quickly utilized by GAOs once introduced into the reactors (Miao et al. 2016). The metabolism of GAOs not only related to energy production and conversion but also produced secondary metabolites (as shown in Figs. 4a, 5a), which could be utilized by denitrifiers as endogenous carbon sources (He et al. 2018b; Lin et al. 2019). The relatively low abundance of AAs hinted that, the metabolism of wheat flour and glucose had scarcely related to oxidation or reduction process. Similarly, the least abundant genes of PLs was attributable to that, wheat flour degradation mainly involved hydrolases, rather than lyases.
Filter clustering based on the above-mentioned level 1 and level 2 of functional categories, i.e., the two different annotation ways, shared a similar pattern (Fig. 6b, c), which was consistent with purification performance shown in Fig. 1. This probably highlighted the importance of carbohydrate metabolism to purification performance in the present filters. It was because the defined purification performance mainly focused on the removal of carbon, nitrogen, and phosphorus. According to the constituents of the raw influent, carbon removal was directly determined by carbohydrate metabolism, while the removal of nitrogen and phosphorus also strongly depended on the sufficiency of carbon sources, such as the process of denitrifying phosphorus removal with organic carbon as electron donors (Lin et al. 2019; Xie et al. 2016). The metabolism of carbohydrates that was closely associated with treatment performance might be ascribed to the low C/N ratio (3.8, Table 1). This also hinted that, carbon shortage was still one of the main factors explaining the low purification performance.
Conclusions
This study tried to decipher the significant differences in purification performance of four different biofilters treating simulating domestic wastewater during start-up from the perspective of microbial characterizations revealed by both 16S rRNA and shotgun metagenomic sequencing. It was found that, filter clustering based on treatment performance, microbial compositions, and metabolic pathways displayed a consistent pattern, suggesting the biological sequestration rather than physical/chemical adsorption was still the main path for pollutant degradation during start-up although the four filters included two new ones. Meanwhile, the mature aerated filter performed the nitrification best. For a typical low C/N ratio domestic wastewater, carbon shortage could be a potent limit for performance improvement. Finally, both structural and functional profilings from shotgun metagenomic sequencing presented a more close association to treatment performance compared to those from 16S rRNA sequencing, suggesting the shotgun metagenomic sequencing might be a better choice in deciphering microbial characterizations.
References
APHA (2005) Standard methods for the examinations of water and wastewater, 21st edn. APHA and AWWA and WEF, Washington
Asshauer KP, Wemheuer B, Daniel R, Meinicke P (2015) Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31(17):2882–2884
Bai YH, Huo Y, Liao KLL, Qu JH (2017) Influence of microbial community diversity and function on pollutant removal in ecological wastewater treatment. Appl Microbiol Biotechnol 101(19):7293–7302
Cai YN, Li D, Liang YW, Luo YH, Zeng HP, Zhang J (2015) Effective start-up biofiltration method for Fe, Mn, and ammonia removal and bacterial community analysis. Bioresour Technol 176:149–155
Cai L, Zheng SW, Shen YJ, Zheng GD, Liu HT, Wu ZY (2018) Complete genome sequence provides insights into the biodrying-related microbial function of Bacillus thermoamylovorans isolated from sewage sludge biodrying material. Bioresour Technol 260:141–149
Cerrone F, Sanchez-Peinado MD, Rodriguez-Diaz M, Gonzalez-Lopez J, Pozo C (2011) PHAs production by strains belonging to Massilia genus from starch. Starch-Starke 63(4):236–240
Chen H, Li A, Wang Q, Cui D, Cui CW, Ma F (2018) Nitrogen removal performance and microbial community of an enhanced multistage A/O biofilm reactor treating low-strength domestic wastewater. Biodegradation 29(3):285–299
Chen JF, Yang YW, Liu YY, Tang MZ, Wang RJ, Zhang C, Jiang J, Jia CX (2019) Bacterial community shift in response to a deep municipal tail wastewater treatment system. Bioresour Technol 281:195–201
Chu BTT, Petrovich ML, Chaudhary A, Wright D, Murphy B, Wells G, Poretsky R (2018) Metagenomics reveals the impact of wastewater treatment plants on the dispersal of microorganisms and genes in aquatic sediments. Appl Environ Microbiol 84(5):e02168
Delforno TP, Lacerda GV, Noronha MF, Sakamoto IK, Varesche MBA, Oliveira VM (2017) Microbial diversity of a full-scale UASB reactor applied to poultry slaughterhouse wastewater treatment: integration of 16S rRNA gene amplicon and shotgun metagenomic sequencing. Microbiologyopen 6(3):12
Du C, Cui CW, Qiu S, Shi SN, Li A, Ma F (2017) Nitrogen removal and microbial community shift in an aerobic denitrification reactor bioaugmented with a Pseudomonas strain for coal-based ethylene glycol industry wastewater treatment. Environ Sci Pollut Res 24(12):11435–11445
Du L, Trinh X, Chen QR, Wang C, Wang HH, Xia X, Zhou QH, Xu D, Wu ZB (2018) Enhancement of microbial nitrogen removal pathway by vegetation in Integrated Vertical-Flow Constructed Wetlands (IVCWs) for treating reclaimed water. Bioresour Technol 249:644–651
Gao L-J, Xiao Y-T, Gao G-D, Ma C (2007) Characteristics of micro-aerobic water treatment technology and its research progress. Ind Water Wastewater 38(2):5–8
Garcia-Ruiz MJ, Maza-Marquez P, Gonzalez-Martinez A, Campos E, Gonzalez-Lopez J, Osorio F (2018) Performance and bacterial community structure in three autotrophic submerged biofilters operated under different conditions. J Chem Technol Biotechnol 93(8):2429–2439
Garrido-Cardenas J, Polo-Lopez M, Oller-Alberola I (2017) Advanced microbial analysis for wastewater quality monitoring: metagenomics trend. Appl Microbiol Biotechnol 101(20):7445–7458
Gu YF, Wei Y, Xiang QJ, Zhao K, Yu XM, Zhang XP, Li CN, Chen Q, Xiao H, Zhang XH (2019) C: N ratio shaped both taxonomic and functional structure of microbial communities in livestock and poultry breeding wastewater treatment reactor. Sci Total Environ 651:625–633
Guan YJ, Jia J, Wu L, Xue X, Zhang G, Wang ZZ (2018) Analysis of bacterial community characteristics, abundance of antibiotics and antibiotic resistance genes along a pollution gradient of Ba river in Xi'an. China. Frontiers in Microbiology 9:3191
He TX, Xie DT, Li ZL, Ni JP, Sun Q (2017) Ammonium stimulates nitrate reduction during simultaneous nitrification and denitrification process by Arthrobacter arilaitensis Y-10. Biores Technol 239:66–73
He FP, Liu D, Zhang L, Zhai JC, Ma Y, Xu YC, Jiang GS, Rong K, Ma JZ (2018a) Metagenomic analysis of captive Amur tiger faecal microbiome. Bmc Vet Res 14:379
He QL, Song Q, Zhang SL, Zhang W, Wang HY (2018b) Simultaneous nitrification, denitrification and phosphorus removal in an aerobic granular sequencing batch reactor with mixed carbon sources: reactor performance, extracellular polymeric substances and microbial successions. Chem Eng J 331:841–849
Jadeja NB, More RP, Purohit HJ, Kapley A (2014) Metagenomic analysis of oxygenases from activated sludge. Bioresour Technol 165:250–256
Jiang HC, Liu GH, Ma YM, Xu XL, Chen JH, Yang YQ, Liu XH, Wang HC (2018) A pilot-scale study on start-up and stable operation of mainstream partial nitrification-anammox biofilter process based on online pH-DO linkage control. Chem Eng J 350:1035–1042
Jin DC, Kong X, Cui BJ, Bai ZH, Zhang HX (2013) Biodegradation of Di-n-butyl phthalate by a newly isolated halotolerant Sphingobium sp. Int J Mol Sci 14(12):24046–24054
Lin ZY, Wang YM, Huang W, Wang JL, Chen L, Zhou J, He Q (2019) Single-stage denitrifying phosphorus removal biofilter utilizing intracellular carbon source for advanced nutrient removal and phosphorus recovery. Bioresour Technol 277:27–36
Miao L, Wang SY, Li BK, Cao TH, Zhang FZ, Wang Z, Peng YZ (2016) Effect of carbon source type on intracellular stored polymers during endogenous denitritation (ED) treating landfill leachate. Water Res 100:405–412
Miao LZ, Wang PF, Hou J, Yao Y, Liu ZL, Liu SQ (2019) Low concentrations of copper oxide nanoparticles alter microbial community structure and function of sediment biofilms. Sci Total Environ 653:705–713
Mitra S, Forster-Fromme K, Damms-Machado A, Scheurenbrand T, Biskup S, Huson DH, Bischoff SC (2013) Analysis of the intestinal microbiota using SOLiD 16S rRNA gene sequencing and SOLiD shotgun sequencing. Bmc Genomics 14:11
Ng C, Tan B, Jiang XT, Gu XQ, Chen HJ, Schmitz BW, Haller L, Charles FR, Zhang T, Gin K (2019) Metagenomic and resistome analysis of a full-scale municipal wastewater treatment plant in Singapore containing membrane bioreactors. Front Microbiol 10:172
Qin XC, Ji MM, Wu XG, Li CJ, Gao YS, Li J, Wu QY, Zhang XJ, Zhang ZJ (2019) Response of treatment performance and microbial community structure to the temporary suspension of an industrial anaerobic bioreactor. Sci Total Environ 646:229–237
Qu JH, Wang HC, Wang KJ, Yu G, Ke B, Yu HQ, Ren HQ, Zheng XC, Li J, Li WW, Gao S, Gong H (2019) Municipal wastewater treatment in China: development history and future perspectives. Front Environ Sci Eng 13(6):88
Rout PR, Bhunia P, Dash RR (2017) Simultaneous removal of nitrogen and phosphorous from domestic wastewater using Bacillus cereus GS-5 strain exhibiting heterotrophic nitrification, aerobic denitrification and denitrifying phosphorous removal. Bioresour Technol 244:484–495
Sarma PM, Duraja P, Deshpande S, Lal B (2010) Degradation of pyrene by an enteric bacterium, Leclercia adecarboxylata PS4040. Biodegradation 21(1):59–69
Tang JY, Bu YQ, Zhang XX, Huang KL, He XW, Ye L, Shan ZJ, Ren HQ (2016) Metagenomic analysis of bacterial community composition and antibiotic resistance genes in a wastewater treatment plant and its receiving surface water. Ecotoxicol Environ Saf 132:260–269
Tao XF, Guo FY, Zhou Q, Hu FL, Xiang H, Xiao GG, Shang D (2019) Bacterial community mapping of the intestinal tract in acute pancreatitis rats based on 16S rDNA gene sequence analysis. Rsc Adv 9(9):5025–5036
Wang HZ, Lou J, Gu HP, Luo XY, Yang L, Wu LS, Liu Y, Wu JJ, Xu JM (2016) Efficient biodegradation of phenanthrene by a novel strain Massilia sp WF1 isolated from a PAH-contaminated soil. Environ Sci Pollut Res 23(13):13378–13388
Wang QB, Yao RD, Yuan Q, Gong H, Xu H, Ali N, Jin ZY, Zuo JE, Wang KJ (2018) Aerobic granules cultivated with simultaneous feeding/draw mode and low-strength wastewater: performance and bacterial community analysis. Bioresour Technol 261:232–239
Wang W, Li Q, Chai WH, Sun CY, Zhang TH, Zhao CH, Yuan Y, Wang XY, Liu HQ, Ye HQ (2019) Lactobacillus paracasei Jlus66 extenuate oxidative stress and inflammation via regulation of intestinal flora in rats with non alcoholic fatty liver disease. Food Sci Nutr 7(8):2636–2646
Xie E, Ding AZ, Zheng L, Dou JF, Anderson B, Huang XL, Jing RT (2016) Screening and characterizing a denitrifying phosphorus-accumulating bacterium isolated from a circular plug-flow reactor. Environ Technol 37(22):2823–2829
Yang L, Ren YX, Liang X, Zhao SQ, Wanga JP, Xia ZH (2015) Nitrogen removal characteristics of a heterotrophic nitrifier Acinetobacter junii YB and its potential application for the treatment of high-strength nitrogenous wastewater. Bioresour Technol 193:227–233
Yang H, Wang J, Lv Z, Tian J, Peng Y, Peng X, Xu X, Song Q, Lv B, Chen Z, Sun Z, Wang Z (2018) Metatranscriptome analysis of the intestinal microorganisms in Pardosa pseudoannulata in response to cadmium stress. Ecotoxicol Environ Saf 159:1–9
Zhang SY, Ban YH, Xu ZY, Cheng J, Li M (2016a) Comparative evaluation of influencing factors on aquaculture wastewater treatment by various constructed wetlands. Ecol Eng 93:221–225
Zhang YM, Ji PF, Wang JF, Zhao FQ (2016b) RiboFR-Seq: a novel approach to linking 16S rRNA amplicon profiles to metagenomes. Nucleic Acids Res 44(10):13
Zhang S-Y, Zhang S-H, Zhang X-L, Wang G-J (2017) Purification efficiency and influencing factors of combined bio-filters for aquaculture wastewater. Environ Sci 38(6):2419–2428
Zhang ZW, Han YX, Xu CY, Ma WC, Han HJ, Zheng MQ, Zhu H, Ma WW (2018) Microbial nitrate removal in biologically enhanced treated coal gasification wastewater of low COD to nitrate ratio by coupling biological denitrification with iron and carbon micro-electrolysis. Bioresour Technol 262:65–73
Zhang L, Shen Z, Fang WK, Gao G (2019a) Composition of bacterial communities in municipal wastewater treatment plant. Sci Total Environ 689:1181–1191
Zhang S, Zhu C, Xia S, Li M (2019b) Impact of different running conditions on performance of biofilters treating secondary effluent during start-up. Bioresour Technol 281:168–178
Zhao Q, Bilal M, Yue SJ, Hu HB, Wang W, Zhang XH (2017) Identification of biphenyl 2, 3-dioxygenase and its catabolic role for phenazine degradation in Sphingobium yanoikuyae B1. J Environ Manag 204:494–501
Zhao RX, Feng J, Liu J, Fu WJ, Li XY, Li B (2019) Deciphering of microbial community and antibiotic resistance genes in activated sludge reactors under high selective pressure of different antibiotics. Water Res 151:388–402
Zou Y, Lin MX, Xiong WG, Wang M, Zhang JX, Wang MZ, Sun YX (2018) Metagenomic insights into the effect of oxytetracycline on microbial structures, functions and functional genes in sediment denitrification. Ecotoxicol Environ Saf 161:85–91
Acknowledgements
This work was financially supported by the Open Research Program from the Agriculture Ministry Key Laboratory of Healthy Freshwater Aquaculture (ZJK201902), the CRSRI Open Research Program (Program SN:CKWV2019765/KY), the Fundamental Research Funds for the Central Universities (WUT: 2019III107CG), the Natural Science Foundation of Hubei Province (2017CFB511) and the National Natural Science Foundation of China (31202034).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, S., Yuan, J. & Guo, W. Metagenomic insights into microbial characterizations in explaining the distinction of biofilter performance during start-up. Biodegradation 31, 183–199 (2020). https://doi.org/10.1007/s10532-020-09902-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-020-09902-1