Abstract
Clonal plants provide a challenge to ecological management and restoration programs. Clonal plants frequently occur as few genets over large spatial scales and may persist for hundreds of years. Hence population structures, effective population sizes and breeding systems of clonal plants are not well understood or managed at scales applicable to non-clonal species. This is a critical issue because clonality is a common trait including among threatened plants and more studies into the management requirements of clonal plant species are required. We investigate the impact of reducing the population size of a highly clonal species and how these impacts can be minimized. Persoonia hindii has a restricted distribution in forested ridges on Newnes Plateau west of Sydney, Australia. We collated data and surveyed populations, undertook assessments of reproductive patterns (flowering; pollination; fruit set and fall), and modelled pollen dispersal among genets. We relate this assessment to planning approaches uninformed by genet structure and reproductive patterns. Small sites likely function as discrete populations, each with low numbers of genets. Genet diversity at this site scale is important as it is associated with increased fruit set. Loss of plants from populations poses a significant risk to population level fitness if genet diversity is reduced. There is an urgent need for more biologically informed approaches for the management of clonal plant species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Clonality is a common plant trait globally, including among threatened plants (Tepedino 2012). Clonality, however, provides a challenge to ecological management and restoration programs. Clonality can impart high levels of fitness (increased opportunities for sexual reproduction—Crone 2001) and some long-lived clonal species appear immortal (living to 10,000 years or more—Ally et al. 2010) relative to our management horizons (typically ~ 100 years—IUCN 2019). However, at these time scales clones will neither track climate (which may reduce local adaptation/fitness—e.g. Gross et al. 2016; James and McDougall 2014) nor expose progeny to selection. Thus, clones may persist, but be maladapted to new/future climate challenges/conditions (Thomas et al. 2004). Clonality is also associated with impaired sexual processes (Vallejo-Marín et al. 2010). The accumulation of somatic mutations by clones (Gross et al. 2012), for example, leads to outcomes such as reduced pollen viability (Ally et al. 2010) or failure of anther dehiscence (Gross and Caddy 2006). Additionally, incompatibility mechanisms which operate to maintain fitness (avoid inbreeding) in genetically diverse populations, conversely may induce reproductive failure where levels of clonality exceed thresholds leading to mate limitation (an Allee effect—see Davis et al. 2004 and Young et al. 2012 for a more complex example). Few management, planning or restoration programs address these critical genetic and reproductive patterns of clonal species (e.g. Broadhurst and Coates 2017; Frankham et al. 2014).
A particularly challenging aspect of managing clonal species is in relation to the loss of plants from a population. Loss of plants (reducing the population size) may involve the loss of genets (genetically distinct plants) or ramets (genetically the same plants—clones) and the outcome of this loss may be strongly dependant upon whether ramets or genets are lost from a population. At the extreme, if an entire population is lost, this may be critical for highly clonal species because clonal species are often highly genetically structured and the genomes present in any given population may be unique (James and McDougall 2014). These genomes may be locally adapted (Sork 2018), potentially most suited for climate adaptation (e.g. Gross et al. 2016) or critical ancestral linages that require conservation (Hooper 1996; James 1982). Additionally, levels of clonality (and hence spatial genetic structure) can vary substantially across a species’ range, implying that impacts will be different among populations (Wang et al. 2018; Roberts et al. 2016; O’Brien et al. 2014). Further, inter-population dispersal (rare but important long-distance pollen or seed dispersal to adjacent populations—e.g. Pearson and Dawson 2005; Barrett 2015) may also be reduced/lost if populations are removed.
In contrast to loss of an entire population, the consequences of removal of only some plants from a population will depend on the number and distribution of clones in the population. Losses may be relatively unimportant if the population is dominated by a single clone, but not always. Levels of floral display may be reduced and this can reduce floral visitation (e.g. Field et al. 2005) potentially critical to crossing among flowers (Barrett 2010) and outcrossing positively correlates with clonality (Vallejo-Marín et al. 2010). Further, a population may be dominated by a single clone but contain some plants with different genomes (Barrett 2015). Loss of these few plants may effectively render the site sterile through mate limitation when incompatibility mechanisms exist (Busch and Schoen 2008). Additionally, if plants are removed from a population and used for translocation, this may induce outbreeding impacts (e.g. Young and Murray 2000). Although generally considered a minor risk in the face of other ongoing processes (Ralls et al. 2018), clonal species may be particularly prone to outbreeding impacts (Vallejo-Marín et al. 2010).
Given to date the limited consideration of the distinct life history, genetic and breeding system patterns of clonal plants in conservation planning, this study was undertaken to investigate how a clonal species may be impacted by a reduction in population size. This is particularly important given that clonality is both a common trait and common among threatened taxa which are often the target of conservation plans which allow for some reduction in population size (Office of Environment and Heritage 2018). We focus upon instances where populations are reduced in size, rather than entirely removed, because the loss of diversity resultant from entire population destruction is relatively easily assessed (e.g. Tierney et al. 2017). Reducing population size may, however, have complex effects which manifest over long time scales. For example, clones may persist in the population, but fitness (capacity to reproduce sexually) may be severely impacted due to a disproportionate increase in mate limitation (The Allee effect—outlined above). Such breeding system impacts are not generally assessed, despite incompatibility being widespread among clonal species. In part this is because a consideration of a range of factors (e.g. population genetic structure; population spatial patterns; pollen dispersal patterns) and how they interact is required to assess breeding system impacts and how these drive non-intuitive outcomes (e.g. Pfab and Witowski 2000). In such instances modelling can provide systematic assessments and novel insights. To undertake this study we therefore selected a highly clonal species, present entirely outside of formal conservation reserves, previously subjected to loss from a number of populations and with potential for further loss. Specifically, in this study we: 1. Relate population genetic structure to assessments of population size, spatial patterns, breeding system and pollen dispersal. 2. Model the impact of variations in population size, genet number and pollen dispersal on pollination success. 3. Based on this aggregate data, recommend improved approaches for the management of clonal plant species.
We undertook this study on Persoonia hindii. Persoonia is a large Australian genus of the Proteaceae and has attracted research attention (e.g. Emery and Offord 2019; Ayre et al. 2009; Cadzow and Carthew 2000; Field et al. 2005; Rymer et al 2005; Wallace et al. 2002; Bernhardt and Weston 1996; Krauss 1994 and references in each). Hence, the genus provides a relatively well studied model for assessing variability in reproductive ecology, which is important in placing studies of clonality into an evolutionary context. As an example, Persoonia mollis subspecies nectens grows in fire-prone vegetation in the same region as Persoonia hindii. P. mollis subsp. nectens is non-clonal, killed by fire, has low levels of genetic structure and recruits extensively by seed post-fire (Ayre et al. 2009). In contrast, recent observations and data show that P. hindii is clonal, resprouts post-fire and does not recruit extensively post-fire (S1—P. hindii monitoring reports 2014–2016). Other genera in the Proteaceae are also known to have mechanisms which drive clonality (e.g. triploidy—James and McDougall 2014; Lynch et al. 1998 or dehiscence failure of anthers—Gross and Caddy 2006) and species such as Grevillea renwickii are remarkably similar in both habit and habitat to Persoonia hindii (James and McDougall 2014).
Method
Study species
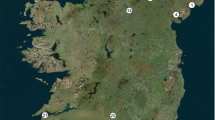
The study species Persoonia hindii is endangered (www.environment.nsw.gov.au/threatenedspeciesapp/profile.aspx?id=10594) and recorded within a relatively restricted area (~ 4700 ha) in public forestry lands on Newnes Plateau west of Sydney (Fig. 1). The species is restricted above the 900 m a.s.l. contour to the highest altitudes of the region (~ 1150 m). Newnes Plateau supports a mix of clay, loam and sandy soils on broad forested ridges intersected by gullies with species rich swamps over a geology predominately sandstone with igneous intrusions (Bembrick 1980). The climate of the study area is cool temperate (mean summer range 12.8–23.3 °C, mean winter range 2.6–9.4 °C; rainfall mean 1885 mm p.a. predominantly in summer at Katoomba—www.bom.gov.au). Persoonia hindii is extensively stoloniferous with stolons observed to extend up to 0.9 m among emergent stems (per obs. of excavations) and genetic data indicates genets may span at least 5 m (Aherns et al. 2020). It also has a clumped distribution pattern with any given site (emergent stems which were within 20 m of another stem were treated as part of a single site) potentially a combination of ramets (emergent stems joined by stolons) and genets (separate genetically distinct plants). Hence survey using a visual estimation of plant numbers is potentially problematic as an estimate of genets. Another particular feature of Persoonia hindii is low flower production. Additionally, while flowers may be produced, there may be no fruit set across an entire site (pers. obs. and S1—P. hindii monitoring reports 2014–16).
Genetic data
Genetic structure within and among sites
Ahrens et al. (2020) assessed genetic patterns among and within 15 sites selected to span the species’ range (Fig. 1). At each of these 15 sites genetic samples were collected from six separate stem clumps using quadrat and dynamic sampling consistent with accepted sampling procedures (details in Ahrens et al. 2020). Samples were analysed using single nucleotide polymorphisms (SNP) via DArTseq™ sequencing (Diversity Arrays Technology Pty Ltd, Canberra, Australia—detail in Ahrens et al. 2020) and returned a pattern of very few genets per site (Table 1).
Genet size
Ahrens et al. (2020) assessed genet size by SNP sequencing (as above) emergent stems within a 25 m2 (5 × 5 m) area in two sites (site 3 from which 49 emergent stems were sampled and site 5 from which 47 emergent stems were sampled—sites shown on Fig. 1). Three genets occurred in each 25 m2 area.
Parentage
Parentage was assessed by sampling and comparing single nucleotide polymorphism among seeds and leaves from three sites (sites 2, 6 and 10—see Ahrens et al. 2020). Mating was non-random, predominantly within a site, dominated by only a few genets and also dominated by selfing (Ahrens et al. 2020).
Site demographics
Abundance and foliage projective cover estimates from offsetting survey
A standard vegetation survey (400 m2) for each of the 15 sites assessed for genetic structure (Fig. 1) was undertaken (March—May 2017) consistent with that mandated for the conservation program of offsetting in the state of New South Wales (Office of Environment and Heritage 2018). This provided a count and foliage projection percent cover estimate for all vascular plants (including Persoonia hindii according to this method) and coincident information about broad habitat features.
Survival and recruitment
Estimates of survival and recruitment were derived from surveys of six sites spread across the species range (sites 1–6; Fig. 1) over three years (May 2014–2016). Sixty-five emergent stem clumps (separated by at least 1 m from other emergent stems) were surveyed. Based on genetic data, these are either independent plants (unconnected ramets or genets) or connected ramets. Hence results are reported per surveyed stem clump and at the site scale. Each clump of emergent stems was tagged and monitored annually for presence (stem survival) or absence (stem mortality). Data on height, number of emergent stems per clump, number of branches, number of flowers and fruits per clump was also recorded (data not reported unless turnover occurred but see S1). Stem clumps that did not survive a year were revisited after another year to confirm no resprouting. Seedling recruitment was also estimated for each of these six sites each May via a 20 min random meander through each site searching for putative seedlings. Minor excavation was used to expose roots to assess if stolons were present from existing ramets.
Fruit set
Across the three-year period in which survival and recruitment were recorded (see above), the 10 monitored stem clumps in site 5 (a small site with only one recorded genet—Table 1) set no fruit. To aid in understanding this pattern, fruit set was then recorded in all 15 sites that were the subject of the survey and genetic analyses. The total number of fruit in each site was estimated monthly from March 2018 until all fruit had fallen (observations had determined that flowering is restricted to January, conversion to fruit occurs by March—with conversion scored when ovules had at least doubled in width compared to ovules in flowers and fruit fall occurs by the December). Fruit were counted individually on each emergent stem if there were less than 50 fruit or if more than 50 fruits occurred an estimate recorded rounded to within 50. Fruit to flower ratios were then calculated from this data.
Comparative population structure and reproductive characteristics
To aid in understanding pollination patterns and to provide alternative site demographic data to the offset survey, further surveys were undertaken of plant density, number of flowering stems and also flower and fruit set. At sites where the pollination study was undertaken (sites 4–6—see below), the number of flowering stem clumps (separate by at least 1 m) and the population structure was measured largely consistent with the method of Rymer (2006) used for other Persoonia. Hence in January 2018 the number of flowering stem clumps (at least 1 m apart) was scored in five randomly chosen 10 m2 plots for each site (these are potentially not independent plants, but this provides data on the spatial distribution of flowers). Flower set was also counted on 20 randomly chosen flowering emergent stem clumps (each at least 1 m apart) at each of the three sites and the height of each of these recorded and averaged. Fruit initiation was also estimated for these sites (see natural pollination levels in “Pollination” section for details). A visual count of all flowering emergent stem clumps at sites was also undertaken to provide an estimate of relative overall site size and flowering and this count was also extended to all 15 sites. Finally, density variability at a fine scale (1 m2 plots) was also measured among all 15 sites by randomly selecting 20 plots within a 10 m2 plot located with one corner randomly chosen to be on the centre marker for each site.
Pollination experiment
Sites 4–6 were used for a pollination study to assess mechanisms driving fruit set-failure (in site 5) and include two adjacent sites from which pollen dispersal might occur. These sites were typical of sites in general; each with few genets, but with a range of emergent stem clump numbers (Table 1). In the third week of December 2017 and 2018 unopened flowers on branches of each of four plants with high levels of flower initiation at each site were bagged using 10 × 15 cm organza bags (www.thepouchplace.com.au) which have a pore size which would exclude pollinators (Neal and Anderson 2004). On each plant a separate branch with unopened flowers was also tagged (the Open Treatment—Treatment A). The Open Treatment flowers were left unbagged and no manipulations were undertaken on flowers for this treatment. Other flowers were inspected weekly and the following treatments (Treatments B, C and D) implemented on flower opening, which commenced in the first week of January. Treatment B—Bagged treatment. Bags were retained on the flowering branch but removed when other pollination treatments (see below) were applied (twice a week) and the stigma of flowers within the bag rubbed with forceps to simulate disturbance (but no pollen applied). Treatment C—Within site cross. A minimum of two flowers with undehisced anthers from each of two plants at least 5 m distant from the experimental plant were selected (a distance over which known pollinators potentially might disperse pollen, but which might include clonal stems or distinct genets). Pollen was extracted from anthers of flowers from one of these donor plants by squeezing and rubbing fine forceps along anthers until a minimum of 50 pollen grains were observed on the forceps (10 × hand lens). Pollen was then applied to stigmas of the previously bagged flowers on a branch of the experimental plant (application success observed with lens). This application was repeated using pollen from donor flowers from different stems and the branch then rebagged (so that all flowers received pollen from two donor flowers from different stems at least 2 m apart within the site). Treatment D—Among site cross. This was undertaken as per the within site cross (above) except pollen was sourced from one of the other sites (Site 6 and 4 were reciprocally pollinated and Site 5 received pollen from Site 6). Treatments were repeated at ~ 2–3 day intervals for 2 weeks. All bags were removed from plants following petal drop (30th January in both years). The number of flowers on each plant that were retained and had initiated fruit development (ovary had expanded to at least twice natural width) was then recorded in the second week of March (both years) and scored as a proportion of total flowers present before pollination.
Natural pollination levels were also assessed in 2018 on five unmanipulated flowering plants in each of the three sites used for the manipulative pollination study. Thus, five plants at each site were selected that were closest to the north, south, east and west boundary and centre of the site. A single branch on each was tagged and number of flowers recorded in the first week of January and these were counted weekly and scored for fruit initiation in the last week of March. Levels of fruit initiation were compared to those of the manipulated pollination treatments.
Pollination experiment analyses
Differences in fruit initiation were assessed using the Anova function in R (version 3.5.2 in RStudio 1.1463—R Development Core Team 2011). Before ANOVA, a test for dispersion difference among treatments was implemented using the cochran.test function in the R package outliers (version 0.14). Where significant dispersion differences were determined (p < 0.05) data was transformed by logit [package gtools version 3.8.1 with all 0 and 1 values converted to 0.025 and 0.975 respectively—see Fox and Sanford (2011) and Warton and Hui (2011)] and then, if required, transformed to binary data to stabilise variance. Following Wassertein et al. (2019), probability values (p) are reported and considered firstly in relation to overall data patterns as well as conventional significance values (p < 0.05). A three-factor ANOVA (factor 1 year = fixed; factor 2 site = fixed; factor 3 treatment with four levels = fixed) was undertaken including all interaction terms among the main factors. A one-factor ANOVA (factor = site; fixed) was undertaken for the natural pollination assessment. Identified differences in factors or interactions were analysed using an SNK test (package agricolae version 1.3–1 in R) to determine where differences lay.
Modelled impacts of variations in ramet number, genet number and pollen dispersal
Model design
Code modified from the program MIGCLIM (Engler et al. 2012) was used for modelling. MIGCLIM implements a cell-based model with cells in this model representing individual plants (Fig. 2). Thus, cells in a grid (iniDist) represent compatible genets (assumed as compatible with all plants). Cells in a second overlaid grid (hsMap) then represent ramets of the dominant clone (self-incompatible). A dispersal function (dispKernel) implements negative exponential dispersal of pollen from compatible genets (Fig. 3). However, dispersal is modified by the parameter barrier. Thus, pollen carryover from compatible genet plants beyond a first encountered dominant clone ramet is reduced by barrier cells—i.e. initially each ramet clone cell was set as a barrier cell (prevents further pollen flow). Randomly reducing barrier cells (i.e. randomly selecting only half ramet cells and then no ramet cells to be barriers) models pollen carryover to second or third ramets (but still with a negative exponential dispersal probability so that very limited dispersal occurs beyond the third ramet cell). Because the spatial relationship of clone ramets and compatible ramets might be important, clone ramet and compatible genet cell locations were randomised (using base R code sample(nrow)) before running replicated (× 100) simulations. Hence three randomisations of ramet and genet cell locations, each followed by 100 replications of the model, generated a mean and standard error for each model variant (see below for variants) of pollen dispersal.
Illustration of the pollen dispersal model where three compatible genets (S, coloured yellow) can disperse pollen to 40 receptive clone plants (C) of with 50% of these also acting as barrier cells (coloured orange) which prevent pollen transfer. Dispersal across cells also decreases as a function of the dispersal kernel (Fig. 3). The arrow shows a potential transfer of pollen with a 0.9, 0.8 and 0.7 probability respectively under high, moderate and low dispersal kernels. If the cell the arrow crossed was a barrier cell (was C and orange) then dispersal would involve crossing three cells (labelled 1, 2 and 3) and dispersal probabilities would be 0.5, 0.4 and 0.2 under high, moderate and low dispersal kernels
Model parameterization
The model was run with eighty-one model variants inclusive of all parameter estimate combinations (Table 2). These scenarios include parameter estimates derived from the field survey, pollination and genetic studies as well as data inferred from related species (details in S2). In sum, genetic data (Ahrens et al. 2020), records for Persoonia hindii and literature on Persoonia pollinators suggest most sites function as discrete populations. Sites range from ~ almost 200 flowering emergent stems to ~ 10 or less (modelled as 180; 90; 45 emergent flowering stems on clone ramets—but extrapolated to consider circumstances where fewer occur). Genetic data (Ahrens et al. 2020) also indicates low genet diversity within each site with a best estimate ranging from 1–8 genets (modelled as 8; 4; 1 compatible genets—see S2). Pollen dispersal (a negative exponential function—Fig. 3) was inferred from a large number of studies (S2) whereby realised pollen dispersal in Persoonia is documented at a scale of metres (model run with dispersal up to 5 grid cells) and pollen carryover likely to few flowering ramet stems (pollen modelled as dispersing across 1–3 flower visitations—S2 for rationale). The model provides a relative assessment of pollen dispersal for a single visitation to each compatible genet to clone ramets at one time.
Model analyses
The importance of each parameter (number of flowering stems; number of genets; dispersal kernel; pollen carryover) contributing to realised pollen dispersal was analysed using a general linear model (routine glm in R stats package 3.5.2 using RStudio Version 0.99.484; R Development Core Team 2011). The model included parameter interactions with significance analysed using the Anova routine in R stats (using chi-square test; p < 0.05; R code in S3). A four-factor Anova was used to analyse the effect of each parameter and also a three-factor Anova (pollen carryover removed) to investigate parameter effects assuming pollinator constancy. Before analysis, homogeneity of variance among models was tested using the routine cochran.test (in R package outliers version 0.14; p < 0.05). Model fit was assessed using the ratio of deviance to null deviance. Deviance is the extent of variation from a perfect model fit and thus null deviance is where the data perfectly fits the model (i.e. 1 = perfect fit, 0 = fit no better than random—code in S3 derived from https://www.bookdown.org/egarpor/PM-UC3M/. Accessed 2 Dec 2019).
Results
Site demographics
Abundance and foliage projective cover estimates from offsetting survey
Abundance of Persoonia hindii within the 400 m2 survey plots across the 15 surveyed sites was 22.6 (± 6.7 se—range 5–100). Foliage projection cover was 2.5% (± 0.63 se). Thus stems on average occupied only a small proportion of the total surveyed area (but with up to 10% cover—site 10 and 5% cover in sites 4 and 5).
Survival and recruitment
Survival across three years (65 emergent stem clumps from 6 sites) was high with stem mortality recorded only in 2016 (2 emergent stems—3%: both unburnt from site 3; one relatively small and one large stem clump). Neither stem clumps had set fruit or resprouted within one year and both were isolated stem clumps and potentially separate ramets or genets. No recruitment was recorded across these six sites over this three-year period.
Fruit set
Putative maturing fruits (ovules ≥ double the width of ovules in flowers) were set across all 15 sites except site 5 (which had more remnant flowers in March than the average number of fruit commencing maturation in other sites, but in site 5 conversion to fruit had failed). Site 14 (a small isolated site of only 5 plant clumps, compared to the 32 plant clumps at site 5) had low fruit set (2 mature fruits which fell in October).
Comparative population structure and reproductive characteristics
The number of flowering clumps measured in five 10 m2 plots at sites 4–6 (7.6 ± 3.0 se) was very high relative to the number of flowering plants recorded for other Persoonia (Table 3). Flower set (36.0 ± 2.5 per flowering emergent stem clump) had little variability across sites—including the putatively sterile site 5. However, this level of flower set was very low compared to flower set on plants of other Persoonia, yet the level of reproductive success of these few flowers was also low as was the flower to fruti ratio relative to other Persoonia species (Table 3). Relative site size of the three sites used for the pollination study as measured by total flowering plant clumps ranged from 32 (site 5) to 171 (site 4). This was within the general overall flowering clump range per site across all 15 sites (39.6 ± 11.0 mean ± se, range 5–171). Percent cover of the three sites used for the pollination study measured in 20 1 m2 plots was slightly elevated relative to that of all 15 sites (3.78 ± 0.12 and 2.34 ± 0.65 mean ± se respectively).
Pollination
Pollination experiment
Data was binary transformed to stabilise variance. ANOVA determined all interactions were not significant. Significant differences were found across the pollination treatment (F3,95 = 8.5421, p = 0.00006), which was a large effect relative to year (p = 0.2609) or site (p = 0.8478)—both statistically non-significant. The SNK test determined the among site and open pollination treatments to have significantly higher fruit initiation than the bagged treatment, while the among site pollination cross was also higher than the within population cross. Thus, the within population cross and bagged treatments both produced very low levels of fruit initiation (which were not significantly different from each other) relative to the open and among population cross (Fig. 4).
Natural pollination
Dispersion differences were relatively small and not significant (Cochran.test p = 0.4414 > 0.05) for natural pollination among sites. Hence ANOVA determined no significant difference among sites (F1,14 = 1.063, p = 0.4524) in fruit initiation which had similar means and large associated variances (Fig. 5). Thus, unmanipulated natural pollination events resulted in levels of fruit initiation (mean = 0.11 among sites) similar to the open treatment in the pollination experiment above (mean = 0.12)—see Figs. 4 and 5.
Modelled impacts of variations in flowering ramet number, genet number and pollen dispersal on realised pollination
The 81 scenario variants returned a range from ~ 0—21 mean realised pollinations with relatively low levels of variance associated with the randomization of genet and ramet locations (Table 2). Therefore, variation in flowering ramet and genet locations were relatively unimportant to modelled pollen dispersal. Overall the model performed strongly based on the assessed deviance ratio (R2 = 0.78). Dispersion differences across the model variants were relatively small and not significant (cochran.test; p = 0.144). The four-factor ANOVA determined a significant three-way interaction of pollen carryover with number of flowering ramets and number of genets (F1,227 = 6.62, p = 0.01). Therefore, pollen carryover, number of ramets and genets acted together to increase the level of pollination. Assuming pollinator constancy (i.e. no overall variation in pollinator behaviour and pollen carryover removed from the ANOVA) the model determined a two-factor interaction among the number of flowering ramets and genets (F1,235 = 6.19, p = 0.014). This means no one variant (ramet, genet or pollen carryover) drove realised dispersal, instead each of these contributes and needs to be considered as potentially affecting pollination level. Yet 14 of the 20 model variants producing less than one realized pollination dispersal had only one genet (models in bold at the bottom of Table 2), while the other seven models had two genets. In contrast, 7 of the 8 model variants that produced more than 10 realised pollen dispersals had 8 genets and none of these had low pollen carryover (models in bold at the top of Table 2).
Discussion
Persoonia hindii is a clonal species distinct in the genus from previously studied species in its population structure and habit. Clones extend over many metres at high density and most sites likely constitute few genets. No recruitment was recorded over a three-year period, including after a fire in three sites which had up to 171 flowering emergent stems (fire is a major driver of recruitment in the fire-prone dry forests in which the species occurs—Gill and Catling 2002). Detailed dedicated studies would be required to investigate genetic turnover through time. However, for this species, visual assessments of population structure have little relationship to measured genet patterns and recorded ‘numbers’ of plants do not reflect genet abundance.
The large extent and likely stability of genet structure in a given site has significant implications for the species’ breeding system. Consistent with previous studies in Persoonia, the experimental pollination treatments in this study determined higher fruit set in Persoonia hindii when crossed among sites with distinct genets, compared to within sites dominated by few related genets. This is potentially driven by either self-incompatibility, genetic load (i.e. inbreeding depression—Cadzow and Carthew 2000) or sterility (Gross et al. 2012). This observed pollen donor distance effect on fruit initiation is consistent with self-incompatibility or genetic load limiting fruit set. That is, among-site pollination significantly increased fruit initiation relative to the within site treatment (including a reciprocal cross which implied pollen and ovules were functional). However, this was among three sites and a diversity of causes may limit fruit set, including somatic mutations, and causes can vary substantially across an entire species range (Gross et al. 2012). This needs careful consideration, for example ex situ collections could be monitored for flower and fruit set to determine if sterile clones may occur which would be of little use in contributing to fitness in re-established populations. More broadly, and despite the above finding, natural pollination initiated fruit set at levels not significantly different to the among site pollination levels. This suggests pollinators are effecting pollination among genets and potentially among sites.
Ahrens et al. (2020) determined most seed to be sired within a site, suggesting limited among-site pollen dispersal. Natural fruit set also occurred in 14 of 15 sites, including isolated sites with few observed emergent stems. Therefore, in most sites, pollinators generally effect some level of fruit set. Native bees are likely natural pollinators of the species, these have been shown to be efficient pollinators across the scale at which ramets occur within sites, but effect lower pollen dispersal at broader scales in other Persoonia (Rymer et al. 2005). The observed ‘natural’ levels of fruit initiation may also have been reduced since the introduction of Apis mellifera, a major floral visitor of some Persoonia (Field et al. 2005). Field assumed Apis mellifera to be a pollinator of the endangered Persoonia bargoensis, but recorded only visitation rather than pollen transfer among flowers. However, Rymer et al. (2005) did record pollen transfer among flowers of a number of Persoonia by Apis mellifera. Apis mellifera relative to native bees, may generally reduce pollen dispersal and outcrossing in Persoonia species (Rymer et al. 2005). However, Apis mellifera is also capable of long-distance pollen dispersal (Beekman and Ratnieks 2000) and the potential also exists for an as yet undetected shift to a two-tailed pattern of pollen dispersal—increased short and long-distance dispersal.
An important finding of this study was that the one isolated site which set no fruit across a three-year period was observed to flower and then did set fruit when hand pollinated by pollen from other sites. Only one genet was recorded in this site, it has a relatively low number of flowering emergent stem clumps (32) and these occur at a high density. We predicted these factors would result in low pollinator service (due to few stem clumps), short pollination dispersal distances (due to high density) and pollination among potentially incompatible ramets (due to only one genet being recorded), relative to most sites. Other factors such as resource limitation (i.e. plants do not produce fruit because they lack the resources to do so) can strongly impact fruit set (Crone and Rapp 2014). However, resource limitation is not consistent with the observation that fruit set occurred in this site when flowers were pollinated by pollen from other sites. This study indicates fruit set is limited by pollination processes, not resource limitation.
It is well established that loss of allelic richness in small populations (loss of allelles at the incompatibility locus/loci) can directly drive population decline via reduced seed production (The Allee effect—e.g. de Santaolalla 2015; Young et al. 2012). This is consistent (but not confirmed) with the results of the pollen experiment. Pollination modelling is also consistent with this effect, with lowest levels of pollen transfer modelled with a single compatible genet. However, ramet and genet numbers and pollination dispersal patterns were also shown to interact in the model, so complex patterns of fruit initiation may occur in this system. Further, mechanisms that drive seed recruitment remain unknown, although very low genet diversity at sites implies seed recruitment is rare. Even if rare, however, seed recruitment may be critical to long-term population fitness if it bolsters genet diversity (Zeng-miao et al. 2015). Additionally, although data in this study is consistent with loss of genetic diversity driving pollen limitation (flowers not pollinated by compatible pollen at levels sufficient to maximize fruit set), resource limitation potentially might also affect fruit maturation and seed production in some sites. Additionally, the pollination model provides a broad assessment of pollination outcomes, but detailed field studies of floral visitation, pollen transfer and pollinator movements within P. hindii populations could potentially provide valuable data to further develop the model.
Overall, loss of genet diversity from sites is considered a potential risk to fitness in Persoonia hindii. This reduction in genotypic diversity at the site level may in effect reduce reproductive capacity, thus site genet patterns must be accounted. Yet conservation planning schemes generally do not assess genet number (e.g. Office of Environment and Heritage 2018), despite the low cost (Appleby et al. 2009) and availability of genetic analyses. This includes previous planning for this species that may have removed up to 1200 ‘plants’ (Erskine et al. 2013).
Clonality is a widespread characteristic (Barrett 2010) which assures persistence independent of reproductive cues (Crone 2001) such as fire regimes. Clonality is also a relatively common trait in threatened taxa globally (Tepedino 2012) and also within threatened taxa in New South Wales where this study was undertaken (in ~ 18% of threatened plants, Department of Planning, Industry and Environment 2019). Such clonal species require a consideration beyond a simple account of observed ‘plants’ that may extend to genetic patterns and breeding system. This does not imply less emphasis or importance should be attached to field survey. Rather, it implies that field survey is required by skilled knowledgeable ecologists who can carefully assess population and species characteristics including a consideration of potential genet patterns, breeding system and the need to undertake genetic analyses. This study highlights the importance of understanding the breeding system and genetic patterns of clonal species. Few if any conservation planning schemes require a considered assessment of how breeding system and genetic patterns potentially interact to drive population fitness.
Based on this study we recommend the following:
-
1.
Levels of clonality of threatened plant species should be routinely assessed and reported in field surveys undertaken for assessing conservation planning impacts (noting that levels of clonality can vary among populations of a species).
-
2.
Impact assessments for threatened clonal plant species need to consider genet diversity and reduction in genet diversity that may result from the removal of plants (both at any given site and across the species), this may require genetic analyses.
-
3.
Plant incompatibility and patterns of observed fruit/seed set be more carefully considered in conservation planning where population size may be reduced. Inbreeding depression may also drive fruit and seed set and should be also considered in these assessments.
The adoption of these recommendations will enable better assessments of population size and reproductive patterns for clonal plant species. These are key parameters for predicting population viability (e.g. Thrall et al. 2014; Traill et al. 2007). However, for clonal species like P. hindii, population size may be poorly inferred from simple survey counts. Similarly, reproductive success (fitness) will not have a simple linear relationship to survey count if incompatibility mechanisms occur. Whilst many long-lived clonal plants may appear immortal and appear to consist of many plants in each population, real population sizes may be small and reproductive success low. More accurately estimating these parameters can be critically important to drive good management decisions for such species.
References
Ahrens C, Tierney DA, Rymer P (2020) Clonality and inbreeding amplifies genetic isolation and mate limitation in a rare montane woody plant (Persoonia hindii; Proteaceae). https://www.biorxiv.org/content/10.1101/2020.05.25.114231v1
Ally D, Ritland K, Otto SP (2010) Aging in a long-lived clonal tree. PLoS Biol 8:e1000454. https://doi.org/10.1371/journal.pbio.1000454
Appleby N, Edwards D, Batley J (2009) New technologies for ultra-high throughput genotyping in plants. Methods Mol Biol 513:19–39
Ayre DJ, Ottewell KM, Krauss SL, Whelan RJ (2009) Genetic structure of seedling cohorts following repeated wildfires in the fire-sensitive shrub Persoonia mollis ssp nectens. J Ecol 97:752–760
Barrett SC (2010) Understanding plant reproductive diversity. Philos Trans R Soc B 365:99–109
Barrett SC (2015) Influences of clonality on plant sexual reproduction. Proc Natl Acad Sci USA 112:8859–8866
Beekman M, Ratnieks FLW (2000) Long-range foraging by the honey-bee, Apis mellifera L. Funct Ecol 14:490–496
Bembrick C (1980) Geology of the Blue Mountains. In: Herbert C, Helby R (eds) Western Sydney basin, a guide to the Sydney basin. Sydney, AU, Geological Survey of NSW, pp 134–161
Bernhardt P, Weston PH (1996) The pollination ecology of Persoonia (Proteaceae) in eastern Australia. Telopea 6:775–804
Broadhurst L, Coates D (2017) Plant conservation in australia: current directions and future challenges. Plant Diversity 39:348–356
Busch JW, Schoen DJ (2008) The evolution of self-incompatibility when mates are limiting. Trends Plant Sci 13:128–136
Cadzow B, Carthew SM (2000) Breeding system and fruit development in Persoonia juniperina (Proteaceae). Cunninghamia 64:941–950
Crone EE (2001) Is survivorship a better fitness surrogate than fecundity? Evolution 55:2611–2614
Crone EE, Rapp JM (2014) Resource depletion, pollen coupling, and the ecology of mast seeding. Ann N Y Acad Sci 1322:21–34
Davis HG, Taylor CM, Lambrinos JG, Strong DR (2004) Pollen limitation causes an Allee effect in a wind-pollinated invasive grass (Spartina alterniflora). Proc Natl Acad Sci 101:13804–13807
de Santaolalla , JLSH (2015) Biología y conservación de las especies de Sonchus sect Pustulati (Asteráceas): endemismos rupícolas del complejo Bético – Rifeño (Mediterráneo Occidental) Phd thesis, Universidad de Sevilla. https://www.researchgatenet/profile/Jose_Silva_Hernandez_De_Santaolalla. Accessed 3 May 2019
Department of Planning, Industry and Environment (2019) Unit of measure outcomes, species final data. Unpublished report, Department of Planning, Industry and Environment, NSW, Australia
Emery NJ, Offord CA (2019) The effects of endocarp, heat-shock and short-term storage on the germination of Persoonia hirsuta (Proteaceae) seeds. Seed Sci Technol 47:107–112
Engler R, Hordijk W, Guisan A (2012) The MIGCLIM R package—seamless integration of dispersal constraints into projections of species distributions. Ecography 35:872–887
Erskine P, Fletcher A, Brownstein G, Blick R (2013) Persoonia hindii: Management and research program. https://data.centennialcoal.com.au/domino/centennialcoal/cc205.nsf/0/BBE73B01E82F7261CA2580D5000C92A5/$file/ATTD8ZNC.pdf
Field D, Ayre DJ, Whelan RJ (2005) The effect of local plant density on pollinator behavior and the breeding system of Persoonia bargoensis (Proteaceae). Int J Plant Sci 166:969–977
Fox J, Sanford W (2011) An R companion to applied regression, 2nd edn. Sage, Thousand Oaks
Frankham R, Bradshaw CJA, Brook BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63
Gill AM, Catling PC (2002) Fire regimes and biodiversity of forested landscapes of southern Australia. In: Bradstock RA, Williams JE, Gill AM (eds) Flammable Australia: the fire regimes and biodiversity of a continent. Cambridge, Cambridge Press, pp 351–369
Gross CL, Caddy HAR (2006) Are differences in breeding mechanisms and fertility among populations contributing to rarity in Grevillea rhizomatosa (Proteaceae)? Am J Bot 93:1791–1799
Gross CL, Nelson PA, Haddadchi A, Fatemi M (2012) Somatic mutations contribute to genotypic diversity in sterile and fertile populations of the threatened shrub, Grevillea rhizomatosa (Proteaceae). Ann Bot 109:331–342
Gross CL, Fatemi M, Simpson IH (2016) Seed provenance for changing climates: early growth traits of nonlocal seed are better adapted to future climate scenarios, but not current field conditions. Restor Ecol 25:577–586
Hopper SD (1996) The use of genetic information in establishing reserves for nature conservation. In: Szaro RC, Johnston DW (eds) Biodiversity in managed landscapes: theory and practice. UK, Oxford, pp 253–260
IUCN (2019) Guidelines for using the IUCN red list categories and criteria, version 14. prepared by the standards and petitions subcommittee. https://www.iucnredlistorg/documents/RedListGuidelinespdf. Accessed 2 May 2019
James SH (1982) The relevance of genetic systems in Isotoma petraea to conservation practice. In: Groves RH, Ride WDL (eds) Species at risk research in Australia. Australian Academy of Science, Canberra, pp 63–71
James EA, McDougall KL (2014) Spatial genetic structure reflects extensive clonality, low genotypic diversity and habitat fragmentation in Grevillea renwickiana (Proteaceae), a rare, sterile shrub from south-eastern Australia. Ann Bot 114:413–423
Krauss SL (1994) Restricted gene flow within the morphologically complex species Persoonia mollis (Proteaceae): contrasting evidence from the mating system and pollen dispersal. Heredity 73:142–154
Lynch AJJ, Barnes RW, Cambecèdes J, Vaillancourt RE (1998) Genetic evidence that Lomatia tasmanica (Proteacae) is an ancient clone. Aust J Bot 46:25–33
Neal PR, Anderson GJ (2004) Does the ‘old bag’ makes a good ‘wind bag’? Comparison of four fabrics commonly used as exclusion bags in studies of pollination and reproductive biology. Ann Bot 93:603–607
O’Brien EK, Denham AJ, Ayre DJ (2014) Patterns of genotypic diversity suggest a long history of clonality and population isolation in the Australian arid zone shrub Acacia carneorum. Plant Ecol 215:55–71
Office of Environment and Heritage (2018) Biodiversity assessment method operational manual—stage 1. Office of Environment and Heritage, Parramatta, NSW, Australia. https://www.environmentnswgovau/topics/animals-and-plants/biodiversity/biobanking. Accessed 2 May 2019
Pearson RG, Dawson TP (2005) Long-distance plant dispersal and habitat fragmentation: identifying conservation targets for spatial landscape planning under climate change. Biol Conserv 123:389–401
Pfab MF, Witkowski ETF (2000) A simple population viability analysis of the critically endangered Euphorbia clivicola RA Dyer under four management scenarios. Biol Conserv 96:263–270
R Development Core Team (2011) R: a language and environment for statistical computing. Austria, Vienna
Ralls K, Ballou JD, Dudash MR, Eldridge MDB, Fenster CB, Lacy RC, Sunnucks P, Frankham R (2018) Call for a paradigm shift in the genetic management of fragmented populations. Conserv Let 11:1–6
Roberts DG, Forrest C, Denham AJ, Ayre DJ (2016) Varying levels of clonality and ploidy create barriers to gene flow and challenges for conservation of an Australian arid-zone ecosystem engineer, Acacia loderi. Biol J Lin Soc 118:330–343
Rymer PD (2006) Plant rarity: species distributional patterns, population genetics, pollination biology and seed dispersal in Persoonia (Proteacae), PhD thesis, School of Biological Sciences, University of Wollongong. https://www.rouoweduau/theses/634/
Rymer PD, Whelan RJ, Ayre DJ, Weston PH, Russell KG (2005) Reproductive success and pollinator effectiveness differ in common and rare Persoonia species (Proteaceae). Biol Conserv 123:521–532
Sork VL (2018) Genomic studies of local adaptation in natural plant populations. J Hered 109:3–15
Tepedino VJ (2012) Overestimating population sizes of rare clonal plants. Conserv Biol 26:945–947
Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collingham YC, Erasmus BFN, de Siqueira MF, Grainger A, Hannah L, Hughes L, Huntley B, van Jaarsveld AS, Midgley GF, Miles L, Ortega-Huerta MA, Peterson AT, Phillips OL, Williams SE (2004) Extinction risk from climate change. Nature 427:145–148
Thrall PH, Encinas-Viso F, Hoebee SE, Young AG (2014) Life history mediates mate limitation and population viability in self-incompatible plant species. Ecol Evolut 4:673–687
Tierney DA, Sommerville KD, Tierney KE, Fatemi M, Gross CL (2017) Trading populations—can biodiversity offsets effectively compensate for population losses? Biodivers Conserv 26:2115–2131
Traill LW, Bradshaw CJA, Brook BW (2007) Minimum viable population size: a meta-analysis of 30 years of published estimates. Biol Conserv 139:159–166
Vallejo-Marin M, Dorken ME, Barrett SCH (2010) The ecological and evolutionary consequences of clonality for plant mating. Annu Rev Ecol Evol Syst 41:191–213
Wallace HM, Maynard GV, Trueman SJ (2002) Insect flower visitors, foraging behaviour and their effectiveness as pollinators of Persoonia virgata R Br (Proteaceae). Aust J Entomol 41:55–59
Wang X, Zhao W, Li L, You J, Ni B, Chen X (2018) Clonal plasticity and diversity facilitates the adaptation of Rhododendron aureum Georgi to alpine environment. PLoS ONE 13:e0197089. https://doi.org/10.1371/journal.pone.0197089
Warton DI, Hui FK (2011) The arcsine is asinine: the analysis of proportions in ecology. Ecology 92:3–10
Wassertein RL, Schirm AL, Lazer NA (2019) Moving to a World Beyond “p < 0.05”. Am Stat 73 Suppl 1:1–19
Young AG, Murray BG (2000) Genetic bottlenecks and dysgenic gene flow in re-established populations of the endangered grassland daisy Rutidosis leptorrhynchoides. Aust J Bot 48:409–416
Young AG, Broadhurst LM, Thrall PH (2012) Non-additive effects of pollen limitation and self-incompatibility reduce plant reproductive success and population viability. Ann Bot 109:643–653
Zeng-miao D, Xin-sheng C, Yong-hong X, Ya-jun X, Zhi-yong H (2015) The role of seedling recruitment from juvenile populations of Carex brevicuspis (Cyperaceae) at the Dongting Lake wetlands China. Sci Rep 5:8646. https://doi.org/10.1038/srep08646
Acknowledgements
This work was supported by research fellowships in the School of Life and Environmental Sciences, The University of Sydney and The Centre for Mined Land Rehabilitation, University of Queensland to David Tierney. The contributions of two anonymous reviewers substantially improved this manuscript. Field assistance was provided by Tom Le Breton, Eren Delgado, Kate Tierney and Heidi Zimmer.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Daniel Sanchez Mata.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tierney, D.A., Ahrens, C., Rymer, P. et al. The interaction of clonality, breeding system and genomics for a highly threatened plant species and the management implications. Biodivers Conserv 29, 3009–3029 (2020). https://doi.org/10.1007/s10531-020-02012-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-020-02012-7