Abstract
Determining the distribution and potential ranges of detrimental invasive species has become an essential task in light of their impacts on the environment. However, this effort has been challenging, especially for global invaders. Our goal was to test whether potential ranges of global invaders can be predicted, and examine the factors that shape them by studying the past, current and potential global distribution of a broad-ranging avian invader. We used the common myna (Acridotheres tristis), one of the most broad-ranging avian invaders whose range is currently expanding globally, as a case study. We collected the first detailed global database of global occurrence (n = 7990) of the common myna over the past 150 years, including records from the native and the introduced ranges. We employed MaxEnt to construct species distribution models (SDM) for the global database using climatic, anthropogenic and environmental factors. We provide evidence that invasive species distributions can be predicted from older records, and that model accuracy requires integrating data from the introduced range. This first comprehensive distribution for an avian invader indicates an extensive expansion in the common myna global distribution, with the potential of large areas worldwide being at risk of common myna invasion, thus threatening local biodiversity globally. Range expansion has been facilitated by proximity to urbanized areas and broad environmental tolerance. Our findings reflect the major role of anthropogenic impact in increasing the global distribution of avian invaders and emphasize the value of using SDMs to inform global management practices.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Growing human impact on native ecosystems has led to a range of effects on biodiversity, including a global rise in the number and range of invasive alien species (McKinney and Lockwood 1999; Meyerson and Mooney 2007; Hulme 2009). Invasive species are often considered to be a primary threat to the environment, leading to the decline of native species and extinctions, negatively affecting human and animal health, jeopardizing food security, and imposing a deleterious effect on the human economy and wellbeing (Lowe et al. 2000; Simberloff 2011; Mori et al. 2018). Understanding the factors that enable successful invasions and delineate introduced ranges is essential to managing the geographic spread of such species (Medley 2010).
The distribution of species is affected by a range of factors, including environmental conditions, the biotic environment and the dispersal ability of the species (Guisan and Thuiller 2005; Soberón 2007). It has been suggested that when these factors are favorable for invasive species, they may also establish due to their lack of natural enemies (Keane and Crawley 2002), high propagule pressure (Lockwood et al. 2009), resource availability (Davis et al. 2000), reproduction intensity (Kolar and Lodge 2001), wide habitat/dietary preferences (Blackburn et al. 2009), broad physiological tolerances (Marchetti et al. 2004), short generation time (Theoharides and Dukes 2007), ability to cope with human proximity (Møller et al. 2015), and high degree of genetic variability (Crawford and Whitney 2010). However, these factors are generally difficult to quantify and reliable data are often scarce, especially in little-studied species, and have therefore been suggested to reduce the accuracy of the correlative models applied in assessing species distribution (Elith 2015). Consequently, environmental conditions is the factor mostly used in delineating or predicting the distribution of a species (Elith 2015).
The use of species distribution models (SDM, sometimes referred to as Environmental Niche Models) has been increasing in recent decades in the areas of ecology and conservation biology (e.g., Elith and Leathwick 2009; Hayes et al. 2015; McCune 2016; Young and Carr 2015), and has recently been used also in invasion biology (Thuiller et al. 2005; Beaumont et al. 2009; Gallien et al. 2012). SDMs are often used for predicting the distribution of species across landscapes, as they allow extrapolation of spatial data in order to produce predictions in areas outside of the sampled geographic extent (Sinclair et al. 2010). This holds great potential for the field of invasion biology, as estimating where species could occur in a certain region may be pertinent in assessing their invasive potential (Elith 2015). In recent years, a growing number of studies have employed SDMs to generate predicted distributions for several species of invasive plants (Beaumont et al. 2009; Gallien et al. 2010), invertebrates (Steiner et al. 2008; Taucare-Ríos et al. 2016), vertebrates (Giovanelli et al. 2008; Buckland et al. 2014), and specifically in birds (Fraser et al. 2015; Crystal-Ornelas et al. 2017).
One of the most common invasive species worldwide is the common myna (Acridotheres tristis), a starling (Sturnid) that occurs naturally in south-east Asia and the Indian subcontinent. The myna has been introduced and subsequently become invasive in every continent except Antarctica (Long 1981; Cramp and Perrins 1994; Forys and Allen 1999; Holzapfel et al. 2006; Saavedra et al. 2015a). It was selected by the IUCN as one of the “100 World’s Worst Invasive Alien Species” (Lowe et al. 2000; Luque et al. 2014), and has been implicated in causing several deleterious ecological changes to the local environment, such as impacting populations of native species by competing for nesting cavities (Grarock et al. 2012; Charter et al. 2016), changing the habitat (Kurdila 1988), and preying on eggs and chicks (Feare 2010; Orchan et al. 2013). This omnivore and generalist species has a wide habitat and dietary range (Cramp and Perrins 1994), and is considered an opportunistic species that frequently forages in human-dominated areas and exploits new feeding opportunities (Sol et al. 2012). Common mynas are mainly sedentary and their average flight distance is relatively short (3 km; Feare and Craig 1999). Common mynas often thrive near humans and are often found in urban or semi-urban areas (Cramp and Perrins 1994; White et al. 2005; Grarock et al. 2014). However, it is currently unclear as to what are the environmental conditions that facilitate their presence on the global scale. Despite being an invasive species of international concern, data pertaining to the current global distribution of the common myna, and its changes over time, are still incomplete. In light of its constant range expansions in newly introduced areas [e.g., in Florida, U.S. (Forys and Allen 1999), in Israel (Holzapfel et al. 2006), and in Spain and Portugal (Saavedra et al. 2015b)], this gap in our knowledge becomes increasingly significant when considering the potential ecological and economic outcomes.
Here we tested whether potential ranges of global invaders can be predicted using the past and current distributions of the common myna, a broad-ranging avian invader. Determining the current distribution range of the common myna enables us to analyze the environmental factors that influence it and predict the potential distribution of this species. Therefore, in this study we generated a global range map of this avian invader and determined the factors that influence it. We also highlight the regions considered more suitable for successful common myna invasion and range expansion. In addition, we test the use of spatially-limited (native-only, introduced-only) and temporally-limited (older, more recent) data-sets in comparison to a comprehensive data-set in order to determine whether they are sufficient for predicting the distribution of an avian invader. This enables us to test whether the current distribution of the common myna can be predicted, by comparing the actual species occurrences with the predictions generated by analyses of limited data-sets. In light of the human-commensal nature of these birds and their exotic origin, we hypothesized that the predicted distribution would include many areas around the globe that are either highly urbanized or are characterized by similar environmental conditions to those in their original range. Determining the environmental factors that enable successful invasion of this species worldwide can help elucidate the mechanism of dispersal of already established birds, potentially reflecting similar predictors of change in the global distribution of ecologically similar species. The results of this study are also expected to aid in identifying high-priority management goals, particularly relating to preventing or limiting the spread of avian invaders by detecting potential suitable habitats.
Methods
Data collection
In order to examine and model the distribution of a global avian invader we collected global occurrence data of common mynas from multiple sources over the past 150 years (a full list of resources is provided in Table S1). The collected records represented both native and alien records, but had not been corrected for eradicated populations, as the latter are more indicative of human efforts than of a natural occurrence. Data was cleaned manually by omitting records that did not contain accurate information on the location of the record (coordinates or a specific location), removing of duplicates and omitting single records that could not be furtherly corroborated by additional sources or unestablished populations, such as in the case of records from Italy (E. Mori, personal communication), France and Germany (GBIF.org, 2015). Initially, the complete data-set comprised 79,923 occurrence records, which were then reduced to 7990 unique records that had been adequately processed (see ‘MaxEnt modelling’). In order to determine the chronological progress in the global distribution of the common myna, we divided the complete set of presence records into four subsets and displayed them spatially as follows: records from the years 1864–1960 (n = 257); until 1980 (n = 405); until 1995 (n = 976); and until 2015 (n = 7990). Since the presence records are spatially filtered so that a minimal distance of 3 km separates between each two records, no account of abundance is reflected in the data-sets (Fig. 1). Additionally, in order to test the accuracy of the model, we divided the data-set into two complementary subsets: the first comprised old records (collected in the years 1864–1960, n = 257, hereafter ‘Old’) versus newer records (collected in the years 1995–2015, n = 6836, hereafter ‘New’); and the second comprised native records (n = 2636, ‘Native’) versus records from introduced populations (n = 5354, ‘Introduced’). This was carried out in order to compare the use of different data-sets for species distribution modeling, since occurrence data are often incomplete. Comparing the subsets from different time periods allowed us to determine whether our models can predict species distribution well into the future using the ‘Old’ subset and whether including only recent records (‘New’) allowed for the inclusion of the historical distribution. In addition, we modeled the complete data-set that comprised all of the records on a contemporary global scale (n = 7990, ‘Total’) in order to compare the results of the partial data-sets with the inclusive data-set.
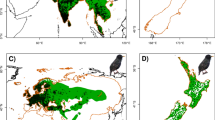
Occurrence records of common mynas distributed globally and filtered (> 3 km) used in this study during four different periods: a until 1960, b until 1980, c until 1995, d until 2015. Area colored grey is the known native range of the species adapted from (Peacock et al. 2007). Presence records are marked with black circles
Environmental variables
In order to describe the factors that influence the distribution of the common myna, we selected a set of 26 environmental variables, of potential biological importance (climatic, topographic, biotic and anthropogenic; Table S3). Variable selection is described in details in the Supplementary Information. We extracted the individual values of every predictor variable for each occurrence from the original variable layer, which ranged in resolution (Table S3). In order to standardize the model sampling, we employed a unified spatial resolution of 0.022 degrees cell size, and obtained a global extent (before selection of background) of (− 49.726455)–58.835963°N and (− 179.253778)–180.072535°E.
In order to reflect the temporal difference between the ‘Old’ data-set and the contemporary ones, it would have been preferable to employ the correct historical environmental variables. However, because of the difficulty of obtaining accurate measurements, and therefore the scarcity of available data-sets, extracting explanatory variables that could also be compared to contemporary models without greatly reducing spatial resolution or excluding important environmental predictors is currently difficult. Therefore, we used the same contemporary resources (Table S3) for the ‘Old’ data-set in our main analyses. However, we performed a similar analysis with historical data for this data-set in order to confirm that the main trends remain (see Supplementary Information for details). The results of this analysis were similar, although of lower accuracy, and reflected the same trends observed in the comparable model presented in the paper.
MaxEnt modelling
We employed the MaxEnt software package (v.3.3.3, Phillips et al. 2006) in order to construct species distribution models. Although MaxEnt was not developed to estimate the distributions of range-shifting species, it has been successfully used for evaluating putative distributions of invasive species (e.g., Giovanelli et al. 2008; Rödder et al. 2008; Ward 2007). However, MaxEnt, like several other modeling algorithms, is known to be sensitive to sampling bias and requires careful spatial filtering (Phillips et al. 2009; Boria et al. 2014).
We constructed all the models with clamping in order to limit feature variation outside of the training data range (Elith et al. 2011), and employed cross-validation as the estimation method of error rate via 100 replicates per model. The smaller subsets (‘Old’ and ‘Native’) were fitted with 25,000 background points created randomly within the selected background in the ‘dismo’ R-package (Hijmans et al. 2017) on R version 3.3.2 (R Core Team 2013), while the number of background points generated for the larger subsets (‘New’, ‘Introduced’ and ‘Total’) was increased to 50,000 (Table 1).
Model tuning comprised primarily three stages; background selection, correcting spatially auto-correlated sampling, and model fitting. Selecting the correct background is crucial for model performance and should be restricted to an area to which the species could have spread (VanDerWal et al. 2009; Jiménez-Valverde et al. 2011b; Elith 2015). Therefore, we selected a 200 km radial buffer zone around each species record based on the largest flight distance documented for the common myna (Parkes and Avarua 2006). This produced a background that allowed for areas with similar conditions to be included in the model, while considering the biological constraints (Figure S1).
We corrected for spatially auto-correlated sampling by applying a filter to the occurrence records, with the minimal distance between records being at least 3 km, a threshold that was selected based on the average foraging distance observed for common mynas (Peneaux and Griffin 2016). This form of spatial filtering accounts for both a bias stemming from higher population densities and for a bias related to a typical increase in human observations reported near anthropogenic centers, and is generally considered sufficient in minimizing sampling bias (Kramer-Schadt et al. 2013; Boria et al. 2014). Accounting for both spatially-biased sampling (background selection) and spatially auto-correlated sampling (spatial filtering) has recently been advocated (Phillips et al. 2017). Percent contribution of each variable was calculated as detailed in Phillips et al. (2006). Finally, we employed the R-package ‘ENMeval’ (Muscarella et al. 2014) in order to optimize model parameters (i.e., regularization multiplier and combination of feature classes) and avoid overfitting for each of the data-sets (Syfert et al. 2013; Boria et al. 2014; Zeng et al. 2016). ‘ENMeval’ executes a set of automated model runs and computes several estimators, yielding a comparable evaluation of all possible models.
We carried out the process of selecting the best model (appropriate predictors, model parameters) in several stages. First, we ran the full model with all 26 environmental variables in ‘ENMeval’ (Muscarella et al. 2014) based on a block design with various regularization multipliers (0.25, 0.50, 1, 1.50, 2, 4, 6) and feature class combinations (L, LQ, H, LQH, LQHP, LQHPT; where L = linear, Q = quadratic, H = hinge, P = product and T = threshold). Second, in order to reduce model complexity, we chose to remove variables that were highly correlated and shown to be of lower significance in variance importance tests carried out by ‘ENMeval’. The preliminary percentage contribution of the different variables was slightly different in each model (Table S4). Therefore, the final predictors set chosen comprised the highest ranking variables that were not highly correlated (Pearson’s |r| < 0.75; Table 1). Following selection of the predictors, we employed ‘ENMeval’ again with the final variables set in a second run of model tuning in a step-wise manner: i.e., with each tuning round, the variable with the lowest contribution importance was removed and the model was run again until only two predictors were left. This was shown to outperform both a priori removal of predictors and not removing highly correlated ones at all (Zeng et al. 2016). Finally, once all the models had been run for each data-set, we chose a final model for each data-set based on the lowest Akaike Information Criterion corrected (AICc) score. The selected model was then run in MaxEnt with the optimal set of predictors, regularization multiplier, and feature class combination using 100 replicates. We generated prediction maps using the model raw output in order to identify areas with habitats suitable for common myna occurrence. Subsequently, we created a binary prediction map by utilizing a threshold-based approach to transform the continuous results into a binary product. Binary projections generated by different thresholds may differ drastically, and choosing the correct threshold may be arbitrary (Norris 2014; Liu et al. 2016). Therefore, we applied five different logistic thresholds: ‘10th percentile training presence’ (‘10th’; Liu et al. 2005); ‘balance training omission, predicted area and threshold value’ as previously described for other invasive species (‘bal’; Giovanelli et al. 2008); ‘minimum training presence’ (‘min’; Phillips et al. 2006); ‘maximum training sensitivity plus specificity’ (‘MAXtr’; Lobo et al. 2008); and ‘equal training sensitivity and specificity’ (‘EQtr’; Liu et al. 2005).
Model evaluation
We utilized the commonly used threshold-independent value of the ‘Area Under the Curve’ (AUC) of the Receiver Operating Characteristic plot to estimate the model’s performance of test data. In addition, we evaluated the accuracy of the model prediction by comparing known localities with the gridded predictions made using model projection, also known as model sensitivity (Hanberry and He 2013). This was calculated as [no. of true positives/total no. of occurrence * 100], where true positives represent occurrences that were located within the predicted range. We evaluated the performance and applicability of the five different thresholds used by comparing sensitivity scores based on each of the thresholds (Lobo et al. 2008).
Results
Past and current global myna distribution
The proportion of records originating from introduced areas increased with time: until 1960, 82.1% of the reported observations were made in the native range, followed by 64.4% until 1980, 39.9% until 1995, and 33.0% out of all the records used in this study (until 2015). Proportionally, since we had obtained the previous records for each data-set and did not omit them from the subsequent one, an increase of 196%, 552%, and 977% can be seen to have taken place in the number of newly reported observations from the introduced areas, respectively.
Model performance
The final models included between eight and eleven explanatory variables for each data-set (Table 1). Model fit as measured by average AUC values was high among models of all of the data-sets and ranged between 0.83 and 0.90, and differences between training AUC (how well the model explains the data used to fit it) and test AUC (how well the model explains independent data) in all data-sets were < 0.001 indicating no overfitting of the model. In contrast, sensitivity scores differed more among the models. On average, the highest sensitivity scores were achieved for the ‘Total’ data-set (Table 1). Conversely, the model based on the ‘Native’ data-set alone resulted in the lowest average sensitivity scores, and in three out of five thresholds received scores below 60 (Table 1). Models based on the ‘Old’ and ‘Introduced’ data-sets achieved intermediate scores (average sensitivity 74.44 < x < 79.32), and the ‘New’ data-set, which included records from both the native and introduced ranges from a recent time period (1995–2015), scored similarly to the complete data-set (85.96).
The different thresholds we applied generated inconsistent results, in which the highest sensitivity value (indicating the percentage of total records that were successfully predicted to occur within a suitable habitat) alternated between data-sets (Table 1). The highest sensitivity scores (93.50–97.64%) were produced by the ‘minimum training presence’ threshold, which was exceedingly nonspecific and predicted species presence in very large parts of the study area that are biologically unlikely to be inhabited by common mynas, such as the Sahara Desert (Figure S2). In contrast, the 10th percentile training presence threshold rendered low sensitivity values for the ‘Native’ (58.42%) and ‘Introduced’ (75.57%) data-sets, but higher ones for the ‘Old’, ‘New’ and ‘Total’ data-sets (92.83%, 86.28%, and 87.80%, respectively). By applying the ‘balance’ threshold, the model based on the ‘Total’ data-set generated the highest sensitivity value (95.33%). Values ranged between 86.42 and 95.20% for the remaining data-sets as well (Table 1). Sensitivity scores based on applying the ‘maximum training sensitivity plus specificity’ and ‘equal training sensitivity and specificity’ thresholds were the lowest and ranged between 44.59–78.04%, and 40.88–75.62, respectively.
The effect of environmental predictors on model output
Variable contribution differed slightly among the five models (Fig. 2). In all five, the factor of ‘impervious surfaces’ was rated highest, ranging from 55.9% to 33.2% (Table S5). Combined with contribution of human density, anthropogenic factors were the most influential in determining myna distribution, followed by temperature-related factors (38.5%–28.5%), especially temperature seasonality (bio4), and precipitation components in the larger data-sets (‘New’—26%, ‘Introduced’—23.8%, ‘Total’—23.7%; Table S5). The probability of common myna occurrence increased rapidly with higher values of both anthropogenic factors, but seemed to have optimal values for temperature and precipitation related predictors, beyond of which the probability of presence decreased (Figure S3). Of the individual temperature components, those related to temperature stability (bio4, which is temperature seasonality or TEMP, interannual temperature variation) had the highest relative contribution in all data-sets and ranged between 13.1 and 32.6% (Table S5). In subsets where precipitation was significant (‘New’, ‘Introduced’, and ‘Total’), precipitation of the driest month (bio14) was the most important singular module (20.5%, 19.1% and 18.5%, respectively), and optimal values ranged between 50 and 150 mm/month. Of the remaining features, none contributed more than 3.6% to explaining common myna presence.
Spatial distribution of predicted suitable habitats
Prediction maps created from all five data-sets indicated that there are multiple locations that are suitable for common myna presence. However, while most data-sets yielded similar suitable distribution for each threshold, the ‘Native’ prediction maps differed from the other data-sets in detecting different areas (Fig. 3). Despite generating either the largest or the second largest predicted distributions, in four out of five thresholds, models based on this data-set scored the poorest or second poorest in model accuracy (Table 1). Additionally, applying different thresholds generated substantially distinct binary prediction maps in all of the data-sets (see Supplementary Information).
Binary maps delineating areas with suitable conditions for common myna presence created by applying the balance training omission, predicted area and threshold on the raw output of MaxEnt model run for the five data-sets used in this study: ‘Old’, ‘New’, ‘Native’, ‘Introduced’ and ‘Total’. Suitable areas for common myna presence are colored dark grey
Because of the effect of the type of data-set and threshold chosen on the generated prediction map, we also provide a continuous map of probabilities of suitable areas for current and future common myna presence based on the ‘Total’ data-set (Fig. 4). This map indicates large areas outside the currently known range of common myna distribution as suitable for the species’ presence, demonstrating that every continent included in the study potentially offers the appropriate environmental conditions for a successful introduction. The map illustrates that the common myna may occur in a wide range of habitats and environmental conditions across a broad latitudinal range.
Discussion
In this study of the common myna we have compiled one of the largest global data-sets for an invasive species ever used in species distribution modeling, spanning 150 years (1864–2015), and incorporating both native and introduced ranges worldwide. We found that using a temporally-limited data-set of old occurrences records (< 1960) was sufficient for adequately predicting the species’ current distribution, thus validating the use of SDMs as a spatial tool in invasive species management. Conversely, models that excluded records from introduced areas did not perform as well. We found a dramatic increase in the current global distribution of the common myna, reflecting extensive introduction events as well as subsequent range expansions. These changes were facilitated by urbanization-related factors and the broad environmental tolerance of this species. Finally, we identified areas outside the current range that may be at risk for common myna invasion and subsequent range expansion. Our findings emphasize the major role of anthropogenic effects in changing the global distribution of a globally successful avian invader.
Here, we provide evidence of global range expansions of an avian invader that occurred in areas in which it was introduced. Introduction events of the common myna have been documented since as early as the mid-nineteenth century across a wide range of areas (Long 1981; Cramp and Perrins 1994). These introductions were either accidental or intentional (Hone 1978; Baker and Moeed 1987), occurred over a continuous period of time, and are still occurring today. Recent introductions include Israel (Holzapfel et al. 2006), Florida (Forys and Allen 1999), Spain and Portugal (Saavedra et al. 2015b), and contain the possibility of secondary range expansions into neighboring countries (Ramadan-jaradi 2011; Khoury and Alshamlih 2015). Currently, there is no evidence of natural range expansions occurring in the native range of the common myna, albeit probably due to scarce documentation, and all of the range expansions reported have been the result of man-made introductions. As human-mediated range expansions are becoming more ubiquitous, natural barriers are becoming less relevant to dispersal capacities (McNeely 2001; Wilson et al. 2009). In the common myna, once an introduction event has taken place and successful establishment has occurred, range expansion then occurs (Fig. 1).
Our results validate the use of SDMs as a tool in invasive species management, as they confirm—with actual presence data collected more than 55 years after the records that were used to fit the model—that even older, outdated data can adequately predict species distributions. We demonstrate this by comparing subsets of the data from different times that included both native and introduced records (‘Old’ and ‘New’). Whereas the sensitivity scores of the model based on a newer data-set were higher in four out of five thresholds (Table 1), the average sensitivity difference calculated across thresholds between ‘Old’ and ‘New’ models was only 10% (74.44% and 85.96% in ‘Old’ and ‘New’ data-sets, respectively). This difference may reflect the size of the data-set (n‘Old’ = 257, n‘New’ = 6836) rather than the time of collection, as previous studies have demonstrated that small sample size may affect model performance (Stockwell and Peterson 2002). Since the ‘Old’ data-set contains records from both native and introduced areas, we believe that its relatively high accuracy stems from an adequate representation of the species and is sufficient for accurately predicting suitable habitats for this species. Furthermore, when comparing the results from the ‘New’ (n = 6836) and ‘Total’ (n = 7990) data-sets, these were highly similar across all thresholds (Table 1), suggesting that when samples size is large enough, a substantial addition of record points may add little to model accuracy. Nevertheless, when using small sample sizes, caution must be taken in interpreting the results, especially when applied to identifying distributions of rare or endangered species, and should be used in conjunction within a broader tool-set or for data exploration (Pearson et al. 2007; Wisz et al. 2008). In the context of invasive species, overestimation of the range limit of a species may pose less of a problem, but should still be used with caution.
Including occurrences from the introduced range in invasive species distribution models is still under debate. Incorporating records from introduced regions may enable the models to include habitat conditions that are suitable for the species but were unavailable to it in its native range, potentially expanding the modeled niche to the fundamental niche (Broennimann and Guisan 2008; Jiménez-Valverde et al. 2011a). The highest average sensitivity scores were obtained for the ‘Total’ data-set, which included data from both the native and the introduced range (sensitivity = 86.96), and which also had a fairly high AUC score (see Supplementary Information). In contrast, the model applied for the data-set that included only occurrences from the native range achieved the lowest scores, while results for the ‘Introduced’ model obtained intermediate scores (Table 1). The difference in the environmental characteristics of the suitable native and introduced habitats may be due to several reasons, including abundant resources and a lack of competitors (Strubbe et al. 2015), historical and geographical constraints (Beaumont et al. 2009; Elith 2015), release from enemies and competitors (Broennimann et al. 2007; Beaumont et al. 2009), and adaptive evolution of the species in its new environment (Broennimann et al. 2007; Beaumont et al. 2009). However, a species in its invasion range may still be spreading and is rarely at equilibrium, thus violating the basic assumption of species distribution models (Elith 2015). While introduced populations of the common myna are still spreading (Fig. 1), our results emphasize the need to incorporate records of invasive species from the introduced range in order to accurately predict suitable potential habitats in non-native regions (Fig. 3, Table 1).
Our prediction models of suitable habitats imply that invasion may occur in many countries (Fig. 4) in the event of a new introduction or through natural expansion from adjacent invasions, as has been the case in Lebanon and Jordan (Ramadan-jaradi 2011; Khoury and Alshamlih 2015). Although common mynas have been introduced into many areas on virtually every continent, few countries have adopted official management measures or undertaken population control (Millett et al. 2004; Parkes and Avarua 2006; Feare 2010; Saavedra et al. 2010; Canning 2011). By generating prediction maps that illustrate potential habitats for common myna presence, we provide precedent knowledge for the appropriate authorities to incorporate during risk assessment of this species. Elith (2015) suggests using such knowledge to target areas susceptible to invasion and improve collaborations among local authorities in order to combat biological invasions. Since the successful eradication of an established population of common mynas is rare and requires continuous efforts (Feare 2010; Saavedra et al. 2010; Canning 2011), such advance information is highly useful when considering what preventive measures to take in order to avoid introduction events altogether.
Elucidating the environmental factors that contribute to an introduced species presence may be of use when assessing other invasive species. Our results indicate that a particular anthropogenic factor (impervious surfaces) is a key influence on the distribution of this species. This echoes the commensal nature of this bird, which is known to thrive near human activities (Cramp and Perrins 1994; Grarock et al. 2014), and may explain its range expansion into areas where natural environmental variables are not optimal (such as more arid areas) by using human settlements as stepping stones into novel environments. This correlation gains additional weight when considering the invasive potential of commensal species, particularly those that exploit human-mediated range expansions (Marambe et al. 2001; Ward et al. 2005; Marzluff and Neatherlin 2006; Møller et al. 2015). In birds, range expansion of commensal species associated with an increase in urbanization was reported in several species (Marzluff et al. 2001; Crooks et al. 2004), and ‘human footprint’ (a quantitative measure of human population size, land use and infrastructure) was an important factor in explaining the establishment success of some alien species (Strubbe and Matthysen 2009; Ancillotto et al. 2016). In common mynas, a strong association with the urban landscape has also been described (White et al. 2005; Peacock et al. 2007; Grarock et al. 2014). Our results indicate that while human density is significantly correlated with common myna prevalence, it is, rather, the level of impervious surfaces that was shown to bear the greatest influence on the species’ current distribution in all five models. We suggest that this can be explained by relating common myna presence to the level of disturbance rather than to the number of people. That is, even in highly disturbed urban landscapes such as industrial areas or infrastructures along major roads where human population density is relatively low, the appropriate conditions for common myna presence are present. This implies that the modification of the landscape in itself affects the distribution of the species, either directly (i.e., by creating artificial nesting cavities) or indirectly (by creating suboptimal conditions for non-synanthropogenic native species).
Additional factors were also found to be correlated with common myna presence in this study, particularly temperature. Components related to temperature stability or range, rather than individual elements such as maximum or minimum temperatures, contributed more to explaining the variance observed in the models (Table S5). This effectively represents the mynas’ tolerance to temperature fluctuations, as these predictors are able to differentiate areas with similar mean temperature but wider minimum–maximum ranges. Optimal values were relatively similar across all data-sets, and indicated that the probability of presence of common mynas is highest in moderately temperature-stable environments (Figure S3). Additionally, in models based on larger data-sets (‘Introduced’, ‘New’ and ‘Total’), precipitation of the driest month was also instrumental in determining the probability of common myna presence, again displaying a moderately flexible scale of their occurrence. These factors indicate that common mynas can occur in a relatively wide range of environments, thus potentially enabling successfully introduced birds to establish in diverse new environments. These findings match previous findings that correlated invasiveness with a broad environmental tolerance (e.g., (Dukes and Mooney 1999; Peterson et al. 2005; Zerebecki and Sorte 2011). The relative flexibility related to minimum environmental precipitation in the common myna may be caused by a broad diet, heavily based on human discards in introduced areas (Machovsky-Capuska et al. 2016), thereby circumventing the need to rely on net primary production which depends greatly on precipitation. A tendency for invasive species to broaden their adaptability to environmental conditions in comparison with their native range has been previously reported (Oduor et al. 2016), and emphasizes the need to incorporate data from the introduced range when predicting potential global distribution.
Creating binary maps has been the final step in many distribution modeling studies, as it is commonly used for various model applications such as delineating range expansions (De Marco et al. 2008), identifying areas susceptible to species invasion (Ward 2007), preliminary detection of suitable areas for biological surveys or reintroductions (Graham et al. 2004) and mapping areas for conservation management (Graham et al. 2004; Waltari and Guralnick 2009). However, binary predictions require an appropriate threshold, which may change due to the purpose of the study, sample size, previous knowledge of species characteristics and life history, and more (Nenzén and Araújo 2011; Norris 2014). Previous studies have obtained fundamentally different results using different thresholds, and supporting empirical data are still scarce (Liu et al. 2005, 2016; Norris 2014). Our results indicate that threshold selection dramatically affects the size and spatial shape of the binary maps generated, and resonate some of the findings previously shown by others (see Supplementary Information).
Carrying out a large-scale global study required us to confront several limitations. While the data-set we compiled is extensive, it reflects a bias in sampling effort, probably towards recent years due to increasing awareness of the significance of data collection, and towards areas that were more accessible to those submitting the reports. However, we feel confident in stating that this is the most comprehensive and updated data-set of common myna occurrence to have been collected and analyzed to date with regard to the distribution of the species. In addition, while SDMs may benefit from more accurate resources (e.g., climatic data), and from including other biologically relevant information (e.g., local biotic interactions), this is difficult to implement in a global study such as the present one (see Supplementary Information). Another important caveat of applying a species distribution model to a species that is in a state of on-going invasion is that it inherently violates the basic assumption of these models, according to which they may only be applied to species that are at quasi-equilibrium with their environment (Guisan and Thuiller 2005; Elith 2015). Since common mynas are clearly still expanding their range in their continental introduced areas (Fig. 1), the reliability of the distributional data (presence records and pseudo-absences) may be questionable. However, in a world of constant change due to rapid urbanization and global warming, few species are at true equilibrium with their environment. We advocate SDM’s as an important tool in the study of the distribution potential of invasive species; and, despite these potential drawbacks, they should be a part of the tool-box for addressing this key subject.
References
Ancillotto L, Strubbe D, Menchetti M, Mori E (2016) An overlooked invader? Ecological niche, invasion success and range dynamics of the Alexandrine parakeet in the invaded range. Biol Invasions 18:583–595. https://doi.org/10.1007/s10530-015-1032-y
Baker AJ, Moeed A (1987) Rapid genetic differentiation and founder effect in colonizing populations of common mynas (Acridotheres tristis). Evolution (N Y) 41:525–538
Beaumont LJ, Gallagher RV, Thuiller W et al (2009) Different climatic envelopes among invasive populations may lead to underestimations of current and future biological invasions. Divers Distrib 15:409–420. https://doi.org/10.1111/j.1472-4642.2008.00547.x
Blackburn TM, Cassey P, Lockwood JL (2009) The role of species traits in the establishment success of exotic birds. Glob Change Biol 15:2852–2860. https://doi.org/10.1111/j.1365-2486.2008.01841.x
Boria RA, Olson LE, Goodman SM, Anderson RP (2014) Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecol Model 275:73–77. https://doi.org/10.1016/j.ecolmodel.2013.12.012
Broennimann O, Guisan A (2008) Predicting current and future biological invasions: both native and invaded ranges matter. Biol Lett 4:585–589
Broennimann O, Treier UA, Müller-Schärer H et al (2007) Evidence of climatic niche shift during biological invasion. Ecol Lett 10:701–709. https://doi.org/10.1111/j.1461-0248.2007.01060.x
Buckland S, Cole NC, Aguirre-Gutiérrez J et al (2014) Ecological effects of the invasive giant madagascar day gecko on endemic Mauritian geckos: applications of binomial-mixture and species distribution models. PLoS ONE. https://doi.org/10.1371/journal.pone.0088798
Canning G (2011) Eradication of the invasive common myna, Acridotheres tristis, from Fregate Island, Seychelles. Phelsuma 19:43–53
Charter M, Izhaki I, Ben Mocha Y, Kark S (2016) Nest-site competition between invasive and native cavity nesting birds and its implication for conservation. J Environ Manag 181:129–134. https://doi.org/10.1016/j.jenvman.2016.06.021
Cramp S, Perrins CM (1994) Handbook of the birds of Europe, the Middle East and Africa. The birds of the western Palearctic vol VIII: crows to finches. Oxford University Press, Oxford
Crawford KM, Whitney KD (2010) Population genetic diversity influences colonization success. Mol Ecol 19:1253–1263. https://doi.org/10.1111/j.1365-294X.2010.04550.x
Crooks KR, Suarez AV, Bolger DT (2004) Avian assemblages along a gradient of urbanization in a highly fragmented landscape. Biol Conserv 115:451–462
Crystal-Ornelas R, Lockwood JL, Cassey P, Hauber ME (2017) The establishment threat of the obligate brood-parasitic Pin-tailed Whydah (Vidua macroura) in North America and the Antilles. Condor 119:449–458. https://doi.org/10.1650/CONDOR-16-150.1
Davis MA, Grime JP, Thompson K et al (2000) Fluctuating resources in plant communities: fluctuating resources a general of invasibility theory. J Ecol 88:528–534. https://doi.org/10.1046/j.1365-2745.2000.00473.x
De Marco P, Diniz-Filho JAF, Bini LM (2008) Spatial analysis improves species distribution modelling during range expansion. Biol Lett 4:577–580
Dukes JS, Mooney HA (1999) Does global change increase the success of biological invaders? Trends Ecol Evol 14:135–139. https://doi.org/10.1016/S0169-5347(98)01554-7
Elith J (2013) Predicting distributions of invasive species. 1–28
Elith J (2015) Predicting distributions of invasive species. In: Walshe TR, Robinson A, Nunn M, Burgman MA (eds) Risk-based decisions for biological threats. Cambridge University Press, Cambridge
Elith J, Leathwick JR (2009) Species distribution models: ecological explanation and prediction across space and time. Annu Rev Ecol Evol Syst 40:677–697. https://doi.org/10.1146/annurev.ecolsys.110308.120159
Elith J, Phillips SJ, Hastie T et al (2011) A statistical explanation of MaxEnt for ecologists. Divers Distrib 17:43–57. https://doi.org/10.1111/j.1472-4642.2010.00725.x
Feare CJ (2010) The use of Starlicide ® in preliminary trials to control invasive common myna Acridotheres tristis populations on St Helena and Ascension islands, Atlantic Ocean. Conserv Evid 7:52–61
Feare C, Craig A (1999) Common myna, Acridotheres tristis. In: Starlings and mynas. Princeton University Press, Princeton, pp 157–160
Forys EA, Allen CR (1999) Biological invasions and deletions: community change in south Florida. Biol Conserv 87:341–347. https://doi.org/10.1016/S0006-3207(98)00073-1
Fraser D, Aguilar G, Nagle W et al (2015) The house crow (Corvus splendens): a threat to New Zealand? ISPRS Int J Geo-Inf 4:725–740. https://doi.org/10.3390/ijgi4020725
Gallien L, Münkemüller T, Albert CH et al (2010) Predicting potential distributions of invasive species: Where to go from here? Divers Distrib 16:331–342
Gallien L, Douzet R, Pratte S et al (2012) Invasive species distribution models—how violating the equilibrium assumption can create new insights. Glob Ecol Biogeogr 21:1126–1136
Giovanelli JGR, Haddad CFB, Alexandrino J (2008) Predicting the potential distribution of the alien invasive American bullfrog (Lithobates catesbeianus) in Brazil. Biol Invasions 10:585–590. https://doi.org/10.1007/s10530-007-9154-5
Graham CH, Ferrier S, Huettman F et al (2004) New developments in museum-based informatics and applications in biodiversity analysis. Trends Ecol Evol 19:497–503
Grarock K, Tidemann CR, Wood J, Lindenmayer DB (2012) Is it Benign or is it a pariah? Empirical evidence for the impact of the common myna (Acridotheres tristis) on Australian birds. PLoS ONE. https://doi.org/10.1371/journal.pone.0040622
Grarock K, Tidemann CR, Wood JT, Lindenmayer DB (2014) Are invasive species drivers of native species decline or passengers of habitat modification? A case study of the impact of the common myna (Acridotheres tristis) on Australian bird species. Austral Ecol 39:106–114. https://doi.org/10.1111/aec.12049
Guisan A, Thuiller W (2005) Predicting species distribution: offering more than simple habitat models. Ecol Lett 8:993–1009. https://doi.org/10.1111/j.1461-0248.2005.00792.x
Hanberry BB, He HS (2013) Prevalence, statistical thresholds, and accuracy assessment for species distribution models. Web Ecol 13:13–19. https://doi.org/10.5194/we-13-13-2013
Hayes MA, Cryan PM, Wunder MB (2015) Seasonally-dynamic presence-only species distribution models for a cryptic migratory bat impacted by wind energy development. PLoS ONE 10:1–20. https://doi.org/10.1371/journal.pone.0132599
Hijmans RJ, Phillips S, Leathwick J, Elith J (2017) dismo: species distribution modeling. R package version 1.1-4. https://CRAN.R-project.org/package=dismo
Holzapfel C, Levin N, Hatzofe O, Kark S (2006) Colonisation of the Middle East by the invasive Common Myna Acridotheres tristis L., with special reference to Israel. Sandgrouse 28:44
Hone J (1978) Introduction and spread of the common myna in New South Wales. Emu-Austral Ornithol 78:227–230
Hulme PE (2009) Trade, transport and trouble: managing invasive species pathways in an era of globalization. J Appl Ecol 46:10–18
Jiménez-Valverde A, Decae AE, Arnedo MA (2011a) Environmental suitability of new reported localities of the funnelweb spider Macrothele calpeiana: an assessment using potential distribution modelling with presence-only techniques. J Biogeogr 38:1213–1223. https://doi.org/10.1111/j.1365-2699.2010.02465.x
Jiménez-Valverde A, Peterson AT, Soberón J et al (2011b) Use of niche models in invasive species risk assessments. Biol Invasions 13:2785–2797. https://doi.org/10.1007/s10530-011-9963-4
Keane RM, Crawley MJ (2002) Exotic plant invasions and the enemy release hypothesis. Trends Ecol Evol 17:164–170
Khoury F, Alshamlih M (2015) First evidence of colonization by common myna Acridotheres tristis in Jordan, 2013–2014. Sandgrouse 37:22–24
Kolar CS, Lodge DM (2001) Progress in invasion biology: predicting invaders. Ecol Evol 16:199–204
Kramer-Schadt S, Niedballa J, Pilgrim JD et al (2013) The importance of correcting for sampling bias in MaxEnt species distribution models. Divers Distrib 19:1366–1379. https://doi.org/10.1111/ddi.12096
Kurdila J (1988) The introduction of exotic species into the United States: there goes the neighborhood. Boston Coll Environ Aff Law Rev 16:95
Liu C, Berry PM, Dawson TP, Pearson RG (2005) Selecting thresholds of occurrence in the prediction of species distributions. Ecography (Cop) 28:385–393. https://doi.org/10.1111/j.0906-7590.2005.03957.x
Liu C, Newell G, White M (2016) On the selection of thresholds for predicting species occurrence with presence-only data. Ecol Evol 6:337–348
Lobo JM, Jiménez-Valverde A, Real R (2008) AUC: a misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr 17:145–151
Lockwood JL, Cassey P, Blackburn TM (2009) The more you introduce the more you get: the role of colonization pressure and propagule pressure in invasion ecology. Divers Distrib 15:904–910. https://doi.org/10.1111/j.1472-4642.2009.00594.x
Long JL (1981) Introduced birds of the world: the worlwide history, distribution and influence of birds introduced to new environments. David and Charles, London
Lowe S, Browne M, Boudjelas S, De Poorter M (2000) 100 of the world’s worst invasive alien species: a selection from the global invasive species database. Invasive Species Specialist Group, Auckland
Luque GM, Bellard C, Bertelsmeier C et al (2014) The 100th of the world’s worst invasive alien species. Biol Invasions 16:981–985. https://doi.org/10.1007/s10530-013-0561-5
Machovsky-Capuska GE, Senior AM, Zantis SP et al (2016) Dietary protein selection in a free-ranging urban population of common myna birds. Behav Ecol 27:219–227. https://doi.org/10.1093/beheco/arv142
Marambe B, Bambaradeniya C, Pushpa Kumara DK, Pallewatta N (2001) The great reshuffling: human dimensions of invasive alien species in Sri Lanka. IUCN, Gland, pp 135–142
Marchetti MP, Moyle PB, Levine R (2004) Invasive species pro ling? Exploring the characteristics of non-native shes across invasion stages in California. Freshw Biol. https://doi.org/10.1111/j.1365-2427.2004.01202.x
Marzluff JM, Neatherlin E (2006) Corvid response to human settlements and campgrounds: causes, consequences, and challenges for conservation. Biol Conserv 130:301–314
Marzluff JM, McGowan KJ, Donnelly R, Knight RL (2001) Causes and consequences of expanding American Crow populations. In: Marzluff JM, Donnelly R (eds) Avian ecology and conservation in an urbanizing world. Springer, Berlin, pp 331–363
McCune JL (2016) Species distribution models predict rare species occurrences despite significant effects of landscape context. J Appl Ecol 53:1871–1879. https://doi.org/10.1111/1365-2664.12702
McKinney ML, Lockwood JL (1999) Biotic homogenization: a few winners replacing many losers in the next mass extinction. Trends Ecol Evol 14:450–453
McNeely JA (2001) The great reshuffling: human dimensions of invasive alien species. IUCN, Gland
Medley KA (2010) Niche shifts during the global invasion of the Asian tiger mosquito, Aedes albopictus Skuse (Culicidae), revealed by reciprocal distribution models. Glob Ecol Biogeogr 19:122–133
Meyerson LA, Mooney HA (2007) Invasive alien species in an era of globalization. Front Ecol Environ 5:199–208
Millett J, Climo G, Shah NJ (2004) Eradication of common mynah Acridotheres tristis populations in the granitic Seychelles: successes, failures and lessons learned. Adv Vertebr Pest Manag 3:169–183
Møller AP, Díaz M, Flensted-Jensen E et al (2015) Urbanized birds have superior establishment success in novel environments. Oecologia 178:943–950. https://doi.org/10.1007/s00442-015-3268-8
Mori E, Meini S, Strubbe D et al (2018) Do alien free-ranging birds affect human health? A global. In: Mazza G, Tricarico E (eds) Invasive species and human health. CABI International Edition, New York, pp 120–129
Muscarella R, Galante PJ, Soley-Guardia M et al (2014) ENMeval: an R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol Evol 5:1198–1205. https://doi.org/10.1111/2041-210X.12261
Nenzén HK, Araújo MB (2011) Choice of threshold alters projections of species range shifts under climate change. Ecol Model 222:3346–3354
Norris D (2014) Model thresholds are more important than presence location type: understanding the distribution of lowland tapir (Tapirus terrestris) in a continuous Atlantic forest of southeast Brazil. Trop Conserv Sci 7:529–547
Oduor AMO, Leimu R, Kleunen M (2016) Invasive plant species are locally adapted just as frequently and at least as strongly as native plant species. J Ecol 104:957–968
Orchan Y, Chiron F, Shwartz A, Kark S (2013) The complex interaction network among multiple invasive bird species in a cavity-nesting community. Biol Invasions 15:429–445. https://doi.org/10.1007/s10530-012-0298-6
Parkes J, Avarua R (2006) Feasibility plan to eradicate common mynas (Acridotheres tristis) from Mangaia Island, Cook Islands. Unpublished Landcare Research Contract Report, Lincoln, New Zealand
Peacock DS, Van Rensburg BJ, Robertson MP (2007) The distribution and spread of the invasive alien common myna, Acridotheres tristis L. (Aves: Sturnidae), in southern Africa. S Afr J Sci 103:465–473
Pearson RG, Raxworthy CJ, Nakamura M, Townsend Peterson A (2007) Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. J Biogeogr 34:102–117. https://doi.org/10.1111/j.1365-2699.2006.01594.x
Peneaux C, Griffin AS (2016) Opportunistic observations of travel distances in common mynas (Acridotheres tristis). Canberra Bird Notes 40:228–234
Peterson MS, Slack WT, Woodley CM, Springs O (2005) The occurrence of non-indigenous Nile tilapia, Oreochromins niloticus (Linnaeus) in coastal Mississippi, USA: ties to aquaculture and thermal effluent. Wetlands 25:112–121. https://doi.org/10.1672/0277-5212(2005)025[0112:TOONNT]2.0.CO;2
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Phillips SJ, Dudík M, Elith J et al (2009) Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecol Appl 19:181–197
Phillips SJ, Anderson RP, Dudík M et al (2017) Opening the black box: an open-source release of Maxent. Ecography (Cop) 40:887–893
Ramadan-jaradi G (2011) Climate variation impact on birds of Lebanon—assessment and identification of main measures to help the birds the adapt to change. Leban Sci J 12:25–32
Rödder D, Solé M, Böhme W (2008) Predicting the potential distributions of two alien invasive Housegeckos (Gekkonidae: Hemidactylus frenatus, Hemidactylus mabouia). North West J Zool 4:236–246
Saavedra S et al (2010) Eradication of invasive mynas from islands. Is it possible. Aliens Invasive Species Bull 29:40–47
Saavedra S, Maraver A, Anadón JD, Tella JL (2015a) A survey of recent introduction events, spread and mitigation efforts of mynas (Acridotheres sp.) in Spain and Portugal. Anim Biodivers Conserv 38:121–128
Saavedra S, Maraver A, Anadón JD, Tella JL (2015b) A survey of recent introduction events, spread and mitigation efforts of mynas (Acridotheres sp.) in Spain and Portugal. Anim Biodivers Conserv 38:121–128
Simberloff D (2011) How common are invasion-induced ecosystem impacts? Biol Invasions 13:1255–1268. https://doi.org/10.1007/s10530-011-9956-3
Sinclair SJ, White MD, Newell GR (2010) How useful are species distribution models for managing biodiverstity under future climates. Ecol Soc 15:8
Soberón J (2007) Grinnellian and Eltonian niches and geographic distributions of species. Ecol Lett 10:1115–1123
Sol D, Bartomeus I, Griffin AS (2012) The paradox of invasion in birds: competitive superiority or ecological opportunism? Oecologia 169:553–564. https://doi.org/10.1007/s00442-011-2203-x
Steiner FM, Schlick-Steiner BC, Vanderwal J et al (2008) Combined modelling of distribution and niche in invasion biology: a case study of two invasive Tetramorium ant species. Divers Distrib 14:538–545. https://doi.org/10.1111/j.1472-4642.2008.00472.x
Stockwell DRB, Peterson AT (2002) Effects of sample size on accuracy of species distribution models. Ecol Model 148:1–13
Strubbe D, Matthysen E (2009) Establishment success of invasive ring-necked and monk parakeets in Europe. J Biogeogr 36:2264–2278. https://doi.org/10.1111/j.1365-2699.2009.02177.x
Strubbe D, Jackson H, Groombridge J, Matthysen E (2015) Invasion success of a global avian invader is explained by within-taxon niche structure and association with humans in the native range. Divers Distrib 21:675–685. https://doi.org/10.1111/ddi.12325
Syfert MM, Smith MJ, Coomes DA (2013) The effects of sampling bias and model complexity on the predictive performance of MaxEnt species distribution models. PLoS ONE. https://doi.org/10.1371/journal.pone.0055158
Taucare-Ríos A, Bizama G, Bustamante RO (2016) Using global and regional species distribution models (SDM) to infer the invasive stage of Latrodectus geometricus (Araneae: Theridiidae) in the Americas. Environ Entomol 45:1379–1385
Theoharides KA, Dukes JS (2007) Plant invasion across space and time: factors affecting nonindigenous species success during four stage of invasion. New Phytol 176:256–273. https://doi.org/10.1111/j.1469-8137.2007.02207.x/pdf
Thuiller W, Richardson DM, Py Ek P et al (2005) Niche-based modelling as a tool for predicting the risk of alien plant invasions at a global scale. Glob Change Biol 11:2234–2250. https://doi.org/10.1111/j.1365-2486.2005.01018.x
VanDerWal J, Shoo LP, Graham C, Williams SE (2009) Selecting pseudo-absence data for presence-only distribution modeling: How far should you stray from what you know? Ecol Model 220:589–594. https://doi.org/10.1016/j.ecolmodel.2008.11.010
Waltari E, Guralnick RP (2009) Ecological niche modelling of montane mammals in the Great Basin, North America: examining past and present connectivity of species across basins and ranges. J Biogeogr 36:148–161
Ward DF (2007) Modelling the potential geographic distribution of invasive ant species in New Zealand. Biol Invasions 9:723–735. https://doi.org/10.1007/s10530-006-9072-y
Ward DF, Harris RJ, Stanley MC (2005) Human-mediated range expansion of argentine ants Linepithema humile (Hymenoptera: Formicidae) in New Zealand. Sociobiology 45:401–407
White JG, Antos MJ, Fitzsimons JA, Palmer GC (2005) Non-uniform bird assemblages in urban environments: the influence of streetscape vegetation. Landsc Urb Plan 71:123–135. https://doi.org/10.1016/j.landurbplan.2004.02.006
Wilson JRU, Dormontt EE, Prentis PJ et al (2009) Something in the way you move: dispersal pathways affect invasion success. Trends Ecol Evol 24:136–144. https://doi.org/10.1016/j.tree.2008.10.007
Wisz MS, Hijmans RJ, Li J et al (2008) Effects of sample size on the performance of species distribution models. Divers Distrib 14:763–773. https://doi.org/10.1111/j.1472-4642.2008.00482.x
Young M, Carr MH (2015) Application of species distribution models to explain and predict the distribution, abundance and assemblage structure of nearshore temperate reef fishes. Divers Distrib 21:1428–1440. https://doi.org/10.1111/ddi.12378
Zeng Y, Low BW, Yeo DCJ (2016) Novel methods to select environmental variables in MaxEnt: a case study using invasive crayfish. Ecol Model 341:5–13. https://doi.org/10.1016/j.ecolmodel.2016.09.019
Zerebecki RA, Sorte CJB (2011) Temperature tolerance and stress proteins as mechanisms of invasive species success. PLoS ONE 6:e14806
Acknowledgements
We thank the associate editor, Adam B. Smith, Emiliano Mori and an anonymous reviewer for their suggestions and comments. We are grateful to Emiliano Mori also for providing additional data for our analyses. We thank Takuya Iwamura, Jonathan Belmaker and Shai Meiri for their helpful comments, and to Adi Barocas for his technical assistance. We thank Naomi Paz for English editing. We also thank Steven Phillips for his vital help. Further thanks are due to Angelo Soto-Centeno for his thoughtful guidance. We are grateful to the Society for Protection of Nature in Israel, Israel Nature and Parks Authority, Shlomit Lifshitz and The Israeli Center for Yardbirds, J. L. Tella and C. Holzapfel for their valuable assistance in records collection.
Funding
This work was supported by the Tel Aviv University Global Research & Training Fellowship in Medical and Life Sciences (GRTF) fund, The Smaller-Winnikow Fellowship Fund for Environmental Research, and The Rieger Foundation-Jewish National Fund fellowship. SK is supported by the Australian Research Council.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Magory Cohen, T., McKinney, M., Kark, S. et al. Global invasion in progress: modeling the past, current and potential global distribution of the common myna. Biol Invasions 21, 1295–1309 (2019). https://doi.org/10.1007/s10530-018-1900-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-018-1900-3