Abstract
The Asian brush-clawed shore crab Hemigrapsus takanoi is a non-indigenous species along the Northern European coast. Although the history of range expansion of European H. takanoi has been well-documented, little is known about the genetic compositions of either the introduced European populations or the native Asian ones. We therefore collected H. takanoi broadly from their native Asian sites and introduced European ranges, and genotyped them by sequencing the mitochondrial 16S RNA gene and by analyzing nuclear microsatellite loci. Our results revealed that the H. takanoi Bay of Seine (France) populations consisted of a genetic admixture between populations in Japan and those in the Yellow Sea region. These French populations should be carefully monitored in the future, since the genetic admixture of multiple source populations may accelerate range expansion in non-indigenous organisms. Our results also suggested that shipping lines from East Asia were more probable vectors than historical juvenile oyster transportations from Japan for the foundation of present European H. takanoi populations. Interestingly, gene flow between populations in Japan and those in the Yellow Sea region (i.e., domestic invasion) was not observed despite the higher potential for artificial translocations via shipping lines in the native Asian range compared with those from Asia to Europe. The lack of domestic invasions implied that intra-specific priority effects of the resident H. takanoi populations played an important role in preventing the successful colonization of artificially-transferred individuals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Evidence of the artificial translocation of organisms beyond their native ranges has become apparent in the last several decades (e.g., Miura 2007; Blakeslee et al. 2010; Cristescu 2015). During the course of artificial translocation, non-indigenous species must experience founding events in the introduced range, and many studies of biological invasions have examined founding events from the standpoint of losing/gaining genetic variations. Specifically, the genetic variation of a founding population of a species would decrease for a genetic bottleneck, while multiple anthropogenic transports from disparate parts of its native range might result in increased genetic variation of the founding population due to genetic admixture (see Miura 2007; Rius et al. 2015). Such genetic admixture may promote invasiveness by increasing the velocity of range expansion following introduction (Simon-Bouhet et al. 2006; Rius et al. 2015) through adaptive evolution and/or heterosis (see Wagner et al. 2017). Thus the comparison of genetic compositions between native and introduced ranges is useful for forecasting the fate of a non-indigenous species.
The Asian brush-clawed shore crab Hemigrapsus takanoi Asakura and Watanabe, 2005 (Brachyura: Varunidae) is a crab species with a native geographical range in East Asia including Far East Russia (Marin 2013), the Korean Peninsula (Lee et al. 2013; Marin 2013) and Japan (Asakura and Watanabe 2005; Mingkid et al. 2006; Yamasaki et al. 2011). Along the Northern European coast H. takanoi is a non-indigenous species (see Asakura and Watanabe 2005): their reproducing population was found for the first time in 1994 along the French Bay of Biscay (Noël et al. 1997). The European H. takanoi population has expanded its range broadly from the Spanish and French Bays of Biscay (Noël et al. 1997) to the German North Sea (Markert et al. 2014). Their range has continued to expand, with populations recently observed in the Kiel Fjord of the Baltic Sea in 2014 (Geburzi et al. 2015) and the southeast coast of Great Britain in 2013–2014 (Wood et al. 2015). Although the history of range expansion of European H. takanoi has been well-documented, little is known about the genetic compositions of either the introduced European populations or the native Asian populations, including the presence or absence of genetic admixture in the introduced range.
It is generally thought that European H. takanoi were likely introduced with Asian oysters and/or by shipping lines (e.g., Noël et al. 1997; Gollasch 1999) possibly via multiple independent introductions into French Atlantic, French British Channel, and the Netherland coast (Markert et al. 2014). Shipping seems to be the most common pathway for the introduction of marine species at a global scale (Molnar et al. 2008; Blakeslee et al. 2017). A recent assessment of the biological invasion risk of marine organisms via global shipping suggested that major international ports in Japan, including those in Tokyo Bay, and in areas around the Yellow Sea were high risk ports for invasions toward Northern European coasts due to heavy traffic (Seebens et al. 2013). Multiple chances of introduction are generally thought to increase the probability of successful colonization of non-indigenous organisms (Katsanevakis et al. 2013; Nunes et al. 2014). Thus shipping may have allowed H. takanoi to establish their populations in Europe from multiple, disparate Asian populations. In the case of Asian oysters, on the other hand, French oyster farms imported juvenile oysters (Crassostrea gigas) from Miyagi Prefecture, Japan (see Koganezawa 1984; Yamamoto 2003; Hatakeyama 2006) back in the 1970s when severe disease killed nearly all European oysters in French aquaculture (Grizel and Héral 1991; Zibrowis 1991). Thus the historical import of juvenile oysters may not have direct links to the range expansion of European H. takanoi in the 1990s and thereafter. Still, if juvenile oyster transfers had been an effective vector for European H. takanoi populations, their source may have been geographically very limited, because juvenile oysters imported to France were largely collected in the Matsushima Bay area, including Mangoku-ura (Koganezawa 1984; Yamamoto 2003; Hatakeyama 2006). In sum, the often-mentioned two possible vectors of European H. takanoi may have produced populations with very different genetic compositions in Europe, but this idea has not been examined to date.
In the present study, we collected H. takanoi broadly from its native range in Japan, Korea and northern China, and from its introduced range in France, the Netherlands and Germany, and genotyped them by sequencing the mitochondrial 16S RNA gene (hereafter mt16S) and analyzing nuclear microsatellite loci. Nuclear microsatellite markers were applied to investigate more contemporary patterns in population history relative to patterns in the mt16S, which generally reflect processes occurring over longer evolutionary timescales. By comparing these data between native and introduced areas, we sought to identify signs of genetic admixture in the introduced European range, and also tried to determine the relative importance of shipping lines and juvenile oyster translocations to the foundation of current European H. takanoi populations. Finally, our data analyses suggested the potential importance of intra-specific priority effects in the context of biological invasions, as also recently suggested by Fraser et al. (2015).
Materials and methods
Sample collection and identification
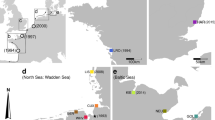
In Japan, 448 individuals of Hemigrapsus takanoi were collected from locations throughout the four major islands over the period from June 2012 to March 2014 (Fig. 1, Supplemental Table S1). Some of these individuals were also included in a previous study (Makino et al. 2015), where detailed information on the sampling methods can be found. From the area around the Yellow Sea, 46 individuals of the Korean H. takanoi population were hand-collected from the intertidal flats during low-tide from three different sites in 2013–2014 (Fig. 1, Supplemental Table S1). The crabs were fixed in 99% ethanol on site. We also purchased 20 individuals of H. takanoi that were collected from an undisclosed coastal area of the Shandong Peninsula in China, i.e., the other side of the Yellow Sea coastal area, and exported to Japan as live fishing bait (see Niwa et al. 2012) in November 2013. The crabs were fixed in 99% ethanol immediately upon arrival. It has been suggested that H. takanoi might be distributed in the southern Japanese islands and Taiwan (see Asakura and Watanabe 2005). Thus we also collected samples from Amami Oshima Island, Japan and Taiwan. As for the introduced European H. takanoi populations, 133 specimens collected from the French Bay of Biscay, the Bay of Seine, the Scheldt Estuary, the Elbe Estuary, Helgoland Island, and Sylt Island (Supplemental Table S1) in 2013–2015 were used. The crabs were fixed in ethanol on site. All of the ethanol-fixed samples were kept at 6 °C until use.
a A neighbor-joining tree showing the relationship between the mt16S haplotypes obtained from Hemigrapsus takanoi and H. penicillatus in the present study, and those obtained from Hemigrapsus crabs on Amami Oshima Island and in Taiwan. Obtained sequences were submitted to DNA Data Bank of Japan under the Accession Nos. LC004189–LC004196 and LC333046–LC333093. The aligned sequences had 464 bp in total, including three 1nt gaps; however, there was no gap-only site. b A map showing the sampling locality in Amami Oshima Island (28.295°N, 129.451°E) and Taiwan (25.082°N, 121.915°E). c Morphological inspections for the Hemigrapsus crabs collected on Amami Oshima Island (red circles) and in Taiwan (red triangles). Panels show relationships between the carapace width and either the relative area of the setal patch on the outer face of male chelae or the spot size on the ventral surface of the cephalothorax in both sexes. Open and closed circles in gray represent data from H. takanoi and H. penicillatus, respectively, collected from the four major Japanese islands (after Makino et al. 2015)
In their native geographical range H. takanoi often coexist with H. penicillatus (Asakura and Watanabe 2005; Yamasaki et al. 2011; Lee et al. 2013; Makino et al. 2015). It has been shown that the mt16S sequence can distinguish these two morphologically-similar species with 100% accuracy (Yamasaki et al. 2011; Markert et al. 2014; see also Supplemental Text 1). Thus we adopted this methodology to guarantee that the specimens examined in the present study were indeed H. takanoi. Specifically, we also sequenced the mt16S of H. penicillatus collected in Matsushima and Tokyo Bays, Japan according to the method mentioned below. These H. penicillatus were also used to characterize the newly developed microsatellite loci, as explained in Supplemental Text 1. The phylogeny of mt16S sequences from both H. penicillatus and H. takanoi (462–464 bp, mostly 463 bp) was illustrated using a neighbor-joining (NJ) tree, which was constructed in MEGA 6.06 (Tamura et al. 2013) by applying Kimura 2 parameter (K2P) pairwise genetic distance and pairwise gap deletions.
Mt16S analysis
Total genomic DNA was extracted individually from muscle tissue using either a DNeasy tissue kit or a Sigma GenElute Mammalian Genomic DNA Miniprep Kit. We then amplified the mt16S gene with the primers HP16s55F and HP16s55R (Yamasaki et al. 2011). Each 15 µL polymerase chain reaction (PCR) cocktail contained 1 µL of DNA template solution, 0.1 µL of EX Taq DNA polymerase (TaKaRa Bio, Otsu, Japan), 1.5 µL of 10X EX Taq buffer, 1.5 µL of dNTP (2.5 mM each), and 0.75 µL of each primer (2.5 µM). The PCR conditions consisted of 1 min of initial denaturation at 94 °C followed by 35 cycles of 30 s at 95 °C, 30 s at 50 °C, and 30 s at 72 °C, with a final extension of 5 min at 72 °C. After cleaning the PCR products with an ExoSap IT kit we carried out cycle-sequencing with a BigDye Terminator sequencing kit. Then sequencing was executed in both the forward and reverse directions with an ABI PRISM 3100-Avant Genetic Analyzer.
We checked all of the mt16S sequences from both directions with FINCH TV (Geospiza). The complete sequences from both directions, which were mostly 463 bp (range 462–464 bp), were aligned with CLUSTAL X (Thompson et al. 1997). There were three mt16S sequences of European H. takanoi, namely Accession Nos. AJ278835 (Schubart et al. 2001), KF982836 and KF982837 (Markert et al. 2014), in the genetic databases. We aligned these haplotypes with those recovered by our own sequencing so as to examine whether there were haplotypes that matched completely with the three previously known European haplotypes. The aligned sequences had 464 bp in total, including three 1-base indels. Using TCS 1.21 (Clement et al. 2000), we also drew an mt16S haplotype network that illustrates all connections having 95% or greater probability of being the most parsimonious, so as to visually represent the relationship among haplotypes recovered from native and introduced areas of H. takanoi.
Haplotype and nucleotide diversities in sampling localities were estimated by using ARLEQUIN 3.5 (Excoffier and Lischer 2010). In order to seek the grouping populations defined as the having the highest differentiation among the groups, analysis of molecular variance (AMOVA) was performed using SAMOVA 2.0 (Dupanloup et al. 2002) without geographical information of the localities. This analysis was conducted separately for (1) exclusively native Asian populations and (2) native Asian and introduced European populations together. In addition, differences in haplotype and nucleotide diversities between the native and introduced populations were statistically treated with a Welch two-sample t test. Furthermore, standardized haplotype richness in native and introduced areas, respectively, was estimated using the rarefaction method implemented in the software ESTIMATES 9.1 (Colwell 2013), and the results were compared to evaluate the reduction of haplotype richness due to bottleneck effects in the introduced area.
Microsatellite analysis
Prior to the present study, we developed 21 new microsatellite markers for H. takanoi and H. penicillatus based on specimens collected in Japan (see Supplemental Text 1). Using 12 of these 21 markers, namely hp01577, hp11192, hp06147, hp13931, hp05535, ht07118, ht19971, ht16728, ht15057, ht14947, ht14226, and ht16035, we conducted microsatellite genotyping in 17 native populations and 8 introduced populations of H. takanoi (Supplemental Table S1).
The native populations included 13 Japanese localities as well as 4 localities of Korea and China around the Yellow Sea (Supplemental Table S1), resulting in genotyping of 299 individuals. The introduced European population included all of the 8 localities, resulting in genotyping of 133 individuals. Using the software package GenAlEX 6.5 (Peakall and Smouse 2012), we calculated on a per-population basis 12 loci-average number of effective (Ne) and private alleles (Np), and expected (He) and observed (Ho) heterozygosity and inbreeding coefficient (F IS), and tested for departure from Hardy–Weinberg equilibrium (HWE).
Using the 12 loci, we evaluated the genetic differentiation among populations by pairwise F ST values with 1023 permutations using Arlequin 3.5. Pairwise F ST values corrected for the presence of null alleles were also estimated with the aid of FreeNA (Chapuis and Estoup 2007). In addition, in order to visualize between-population differentiation in our microsatellite analysis, we performed a discriminant analysis of principal components (DAPC; Jombart et al. 2010), which extracts information from data by first performing a principal component analysis (PCA) on user-defined populations, and then using the PCA factors as variables for a discriminant analysis (DA) to maximize the inter-population component of variation. The analysis was conducted by using the “adegenet” package (Jombart 2008) for the statistical platform R (R Core Team 2015). Differences in mean numbers of per-population basis Ne, Np, and He between the native and introduced populations were statistically examined with a Welch two-sample t-test. Also, the standardized allelic richness in both the native and the introduced areas was estimated for each of the 12 loci; to test the reduction of richness in the introduced area due to bottleneck effects, the rarefaction method implemented in the software ADZE was used (Szpiech et al. 2008).
Results
Native Asian populations
The mt16S sequences clearly separated H. takanoi from the morphologically-similar H. penicillatus (Fig. 1a and Supplemental Text 1). The mt16S haplotypes obtained from Hemigrapsus crabs on Amami Oshima Island and Taiwan Island (Fig. 1b) were very different from those of not only H. takanoi but also H. penicillatus (Fig. 1a). Based on morphological inspections using the relative size of the setal patch on the outer face of male chelae (Takano et al. 1997), these Hemigrapsus crabs were assigned to H. takanoi (Fig. 2c and Table 2); however, inspections based on the spot size on the ventral surface of the cephalothorax (Makino et al. 2015) assigned the Taiwan specimens to H. penicillatus (Fig. 1c and Table 2). Thus the taxonomy of specimens collected in Amami Oshima Island and Taiwan is currently unclear, and in the present study these specimens are collectively referred to as Hemigrapsus spp.
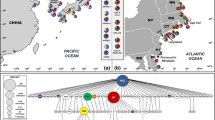
Pie charts representing the frequency distribution of mitochondrial mt16S haplotypes for H. takanoi populations in their native Asian and introduced European ranges. The locality code in Table 1 is shown inside the chart. The number in parenthesis after the locality name denotes the number of individuals sequenced at each sampling site. The colors in the pie charts are identical to those in the mt16S haplotype network, where the area of the circle corresponds to the number of individuals that possessed the haplotype and the smallest circle corresponds to one individual. Missing intermediate haplotypes are colored in black. Also included in the map is information on the mt16S haplotypes of H. takanoi recovered previously from France (Accession No. 278835, deposited as H. penicillatus; Schubart et al. 2001) and Germany (Accession Nos. KF982836 and KF982837; Markert et al. 2014). In addition, the haplotype HTAC002 from continental Asia matched the mt16S sequence of H. takanoi from China (sensu Markert et al. 2014) with Accession No. GU731424 (Xu unpublished)
H. takanoi sensu stricto was distributed in the four major islands of Japan and the area around the Yellow Sea on the Asian Continent, and we recovered 44 mt16S haplotypes from 514 individuals of these H. takanoi (Fig. 1a). All of the mt16S haplotypes were close to each other as shown in the haplotype network; however, there was no shared haplotype between the Japanese H. takanoi population and that of Korea and China, i.e., around the Yellow Sea (Fig. 2). In the latter region, the haplotype HTAC002 (pale green in Fig. 2) was numerically dominant in all 4 localities, and was possessed by 53 of 66 individuals examined. In Japan, the haplotypes HTJP002, HTJP001 and HTJP005 (dark red, dark blue and gray, respectively, in Fig. 2) were numerically dominant and possessed by 62, 25 and 4% of individuals examined there, respectively. While the haplotype HTJP002 was found in all of the Japanese localities examined, the haplotype HTJP001 was mostly found along the Pacific coast of Honshu Island. The haplotype HTJP005 was found mainly on the northern part of Honshu Island including the Pacific coastal areas of Miyagi Prefecture. There was no significant difference either in the haplotype diversity or nucleotide diversity (Table 1) between H. takanoi populations in Japan and the area around the Yellow Sea (Welch two sample t-test, P > 0.05). According to AMOVA, the results of grouping where genetic differentiation among the groups became maximal indicated a combination of four groups, which separated populations around the Yellow Sea (locality code 33–36) as one group from populations in Japan that were divided into three groups (Table 3).
Microsatellite analysis was conducted only for H. takanoi sensu stricto collected in the four major islands of Japan and the area around the Yellow Sea (Table 1). Pairwise F ST values, both with and without considering the effects of null allele presence, tended to be high between populations in Japan excluding Hokkaido Island and populations in Hokkaido Island plus the area around the Yellow Sea (Table S2). In the DAPC analysis, according to the scatter plot of the first two components of DA, the first axis separated populations in Japan excluding Hokkaido Island from populations in Hokkaido Island (locality code 1, 2, and 5) plus those in the area around the Yellow Sea (Fig. 3a). The second axis of DA seemed to further separate populations in Hokkaido Island from those in the area around the Yellow Sea.
Plots of the first two axes obtained by Discriminant analysis of Principal Components (DAPC) of H. takanoi populations in a the native Asian range and b both the native and introduced European ranges. Analyses were conducted with data on the 12 microsatellite loci. To avoid putting too much information in a panel, the same analytical results are drawn in the right and left panels, where individuals of each population are represented with a population-specific symbol (see the legend between the panels). In the right panels, locality codes (see Table 1 and Fig. 2) are placed at the center of each population, and a minimum spanning tree based on the distances between populations within the entire space is shown. In the left panels, individuals of each population are connected with vectors from the center of the population, and populations are delineated by inertia ellipses. Further, eigenvalues of the PCA and DA are displayed in the insets
Introduced European populations, and comparisons with the native populations
From the 133 European H. takanoi specimens, only four mt16S haplotypes, namely HTJP001, HTJP002, HTJP005 and HTAC002, were recovered (Fig. 2, Supplemental Table S1). Thus all of these specimens were H. takanoi sensu stricto, and the haplotypes recovered from Hemigrapsus spp. on Amami Oshima Island and Taiwan were not found in Europe at all.
The mt16S haplotype HTAC002, i.e., the most dominant one in the area around the Yellow Sea, was found only in the two populations along the Bay of Seine, France, where all four haplotypes mentioned above were found. The population in the French Bay of Biscay possessed three Japanese haplotypes, while only HTJP001 and HTJP002 were found from populations in the Netherlands and Germany. According to an AMOVA that searched for the grouping having the maximal genetic differentiation among groups (Table 3), the Bay of Seine populations (locality code 42–43) were suggested as an independent group while other European populations (locality code 37–41 and 44) were merged into one of the groups of the Japanese populations. In the microsatellite analysis, the average pairwise F ST values related to the Bay of Seine H. takanoi populations [shaded in grey in Table S2, 0.062 ± 0.017 (SD) or 0.057 ± 0.018 without or with consideration for the effects of null allele presence, respectively] were ca. three-fold higher than the average value for the rest of the population pairs (0.027 ± 0.015 or 0.023 ± 0.016 without or with consideration for the effects of null allele presence, respectively). In the DAPC analysis, the Bay of Seine populations were clearly separated from the rest of the populations by the first two DA axes (Fig. 3b).
We statistically compared genetic numerical variables (Table 1) between native and introduced H. takanoi populations. Differences neither in mt16S haplotype diversity nor nucleotide diversity were statistically significant, while in the microsatellite genotyping the number of effective and private alleles, and expected heterozygosity were significantly smaller in the introduced populations compares with the native populations (Table S3). The rarefaction analysis showed that the standardized numbers of both mt16S haplotypes and alleles among the 12 microsatellite loci in the European H. takanoi population were smaller than those in the native populations (Fig. 4; see also Table S4).
Discussion
Evidence of multiple discrete sources for European H. takanoi populations, and possible explanations for the geographic structure in native East Asia
We genotyped both native and introduced H. takanoi populations using mitochondrial DNA sequences and nuclear microsatellite loci. In their native area, we surveyed two potential donor regions of European H. takanoi, namely Japan and the area around the Yellow Sea on continental Asia. There was no shared mt16S haplotype in H. takanoi between Japan and the area around the Yellow Sea. Our microsatellite genotyping also revealed the presence of genetic discontinuity between the two donor areas, especially between the Honshu, Shikoku and Kyushu H. takanoi populations and those around the Yellow Sea. The population differentiation based on our microsatellite data was also visualized using a Bayesian clustering algorithm implemented in the software package STRUCTURE 2.3.3 (Pritchard et al. 2000; Supplemental Text 2), and the obtained result was very similar to that of DAPC. Therefore, our results suggest that there is little gene flow, and thus little genetic admixture, across the East China Sea and the Tsushima Straits. Interestingly, in the Bay of Seine H. takanoi populations, we found mt16S haplotypes that were numerically dominant in Japan as well as in the area around the Yellow Sea, implying the sympatric distribution of individuals/descendants from these different native areas. Given that there is little gene flow across the East China Sea and the Tsushima Straits in native H. takanoi populations, our results from the Bay of Seine clearly demonstrate that the current European H. takanoi populations were at least partly formed by multiple introductions from genetically-differentiated, discrete native populations.
We could consider that the genetic differentiations in H. takanoi populations between Japan and the area around the Yellow Sea may have emerged via events on a geographical/evolutionary time scale. Indeed, oscillatory changes in the marine environments would have been severe in East Asia, especially in locations between Continental Asia and Japan, where the Japan Sea is currently, because of glacial–interglacial cycles following the Pleistocene. Specifically, the Japan Sea, which is connected to other seas through shallow and narrow straits, might have been isolated from other surrounding seas for regression during glacial periods, and its salinity might have been drastically decreased by a massive influx of freshwater from Continental Asia (e.g., Ohba et al. 1991; Tada 1994). H. takanoi populations would have been evacuated from the Japan Sea during such suboptimum periods of time. It could therefore be possible that the range of H. takanoi was severely contracted and fragmented during the glacial periods compared with the current range, and that the present genetic structure of H. takanoi in the native area still holds the pattern of historical habitat contractions.
The H. takanoi populations on Hokkaido were somewhat exceptional compared with other Japanese populations, because the microsatellite genotyping suggested a higher similarity with populations around the Yellow Sea rather than geographically-close populations on the major islands of Japan. Note, however, that there was no shared mt16S haplotype between Hokkaido H. takanoi populations and those around the Yellow Sea. If there was any contemporary gene flow, there should also have been shared mt16S haplotypes; otherwise we would need to hypothesize sex-specific differences in colonization success (i.e., zero for females), which would be difficult to accept. Note that there is a possibility that H. takanoi was not distributed in Hokkaido during the glacial periods, not only because the salinity of the Japan sea would have been very low but also because sea temperature in the northern area would have been lower than the current temperature. If these possibilities were the case, we could further consider that, in the early expansion stage of fragmented H. takanoi populations following the recovery of environmental conditions, introgressive hybridization between the Japanese population and Continental Asian population could have occurred. The current H. takanoi Hokkaido populations might have been a descendant from such introgressive hybridization.
Importance of shipping lines as invasion vectors
Since the 1997 report of Noël et al., it has generally been thought that H. takanoi populations were introduced with Asian oysters and/or by shipping lines into Europe. In the case of Asian oysters, juvenile oysters attached to scallop shells were carried airborne from Japan to France without soaking in seawater (Koganezawa 1984; Yamamoto 2003). This process would have enabled the juvenile and adult individuals of H. takanoi to be transferred, rather than their planktonic larvae. Thus, the founder H. takanoi population may have had a rapid start in increasing their population size. If so, European scientists may have found this alien species much earlier than 1993–1994 (Noël et al. 1997; Gollasch 1999), because juvenile oyster transportations to France peaked in 1972 (Koganezawa 1984; Yamamoto 2003). It is also noted that the juvenile oyster transfer eventually stopped in 1980 (Koganezawa 1984; Yamamoto 2003). These pieces of circumstantial evidence suggest that the juvenile oyster translocations cannot be a synchronous vector with the spread of H. takanoi in Europe over the last two decades. As explained below, the present study also found no evidence strongly supporting the idea that the introduction of oysters played an “indispensable” role for the current European H. takanoi populations.
A large portion of the juvenile oysters exported to France in the 1970s were collected in Miyagi Prefecture, especially the area around Matsushima Bay (locality code 9–10; Koganezawa 1984; Yamamoto 2003; Hatakeyama 2006). If the current European H. takanoi populations had been founded mainly by this route, their genetic compositions would be expected to strongly reflect those of populations in the area around Matsushima Bay. Our results, especially the mt16S data (Table 3) on European H. takanoi (excluding the Bay of Seine populations), seem to support this idea. Unfortunately, however, the mt16S data were not without some noise, since the related group in Japan (Table 3) included localities other than the area around Matsushima Bay, such as Tokyo Bay (locality code 14). Our microsatellite data were also not able to separate the Tokyo Bay populations from those in the area around the Matsushima Bay (Fig. 3a, Supplemental Text 2). Taken altogether, therefore, these results do not provide definitive evidence of an indispensable role of juvenile oyster transportation.
It is important that H. takanoi populations in the area around the Yellow Sea were one of the sources of H. takanoi populations in the Bay of Seine, because there is no solid record showing that live oysters were shipped from the area around the Yellow Sea to Europe. Rather, according to Seebens et al. (2013), some of the ports in the area around the Yellow Sea present a high risk for the invasion of marine organisms to Northern European coasts via heavy shipping. Their estimates (Seebens et al. 2013) focus on the transportation of organisms in ballast water, which is effective at transporting invertebrate larvae having a relatively long larval period (e.g., several weeks; see Rius et al. 2015), such as those of H. takanoi, which are thought to have a larval duration of 1 month (Okamoto and Kurihara 1987). In addition to ballast water, transportation via the hulls of ships (Gollasch 1999) and sea-chests (Coutts et al. 2003; Coutts and Dodgshun 2007; Frey et al. 2014) could also be factors. This leads us to conclude that shipping lines were indeed effective vectors for the current H. takanoi populations in the Bay of Seine.
According to Seebens et al. (2013), major international ports in Japan such as those in Tokyo Bay were also high risk ports for invasions toward Northern European coasts due to heavy traffic. Back in 1993, Gollasch (1999) found live H. takanoi in hull-fouling on the automobile-carrying ship SPICA when the vessel was docked at the port of Bremerhaven, Germany. On its way from Asia to Europe, interestingly, SPICA docked at both Japanese ports, including those in Tokyo Bay, and Korean ports (see Gollasch 1999), implying the possibility that a single ship departing from East Asia received propagules from multiple ports across the native range of H. takanoi. As mentioned above, the present genetic analyses did not separate H. takanoi populations in Tokyo Bay (representative of the shipping line hypothesis) from those in the area around Matsushima Bay (representative of the Asian oyster hypothesis); however, there are at least pieces of circumstantial evidence suggesting direct links to the range expansion of European H. takanoi in the 1990s and thereafter in the shipping line hypothesis. Thus we argue that shipping lines from Japan (presumably Tokyo Bay) were also effective vectors for the current H. takanoi populations in not only the Bay of Seine but also other European localities investigated in the present study.
Evidence of genetic admixture of discrete source populations in the introduced European range
In the introduced European range, our rarefaction analyses revealed a reduced genetic variability in the H. takanoi population compared with the native area, probably due to the effects of genetic bottleneck during the course of artificial translocation (Miura 2007; Dlugosch and Parker 2008). As for the genetic composition of each European H. takanoi population, we obtained two kinds of results. In the Bay of Seine populations, on the one hand, we found mt16S haplotypes that were numerically dominant in Japan as well as in the area around the Yellow Sea, implying the sympatric distribution of individuals/descendants from these different native areas. In the remainder of the European H. takanoi populations, on the other hand, we found mt16S haplotypes that were recovered only in Japan. Then, our DAPC analysis (and a supplemental STRUCTURE analysis as well; see Supplemental Text 2) suggested that while the allelic compositions of European H. takanoi populations excluding the Bay of Seine ones were similar to those of Japanese populations, the allelic compositions of the Bay of Seine H. takanoi populations were very different not only from those of the Japanese populations but also from those of populations in the area around the Yellow Sea. Based on these results, we consider that individuals of H. takanoi from Japan and those from the area around the Yellow Sea may have indeed been interbreeding in the Bay of Seine, resulting in the “different” genetic compositions in terms of both mt16S and the 12 microsatellite loci. Thus we argue that genetic admixture of H. takanoi populations across the East China Sea and the Tsushima Straits has been realized in the introduced European range.
Possible mechanisms for the lack of genetic admixture across the East China Sea and the Tsushima Straits: the role of “blocking” effects
International trading via shipping has been intensive not only between European and Asian countries but also across Asian countries, i.e. across the native ranges of H. takanoi. There are at least three pieces of evidence suggesting that the chance of artificial translocation via shipping may be rather higher across the native East Asian ranges of H. takanoi, i.e., domestic invasion (e.g., Hudson et al. 2016), compared with the case from East Asia to Europe. The first piece of evidence is based on the balance of ballast water, which is one of the most notorious vectors for marine non-indigenous species (e.g., Carlton and Geller 1993). Omura et al. (2014) estimated the volume of ballast water transferred to/from 23 major international ports in Japan (mostly in the Honshu and Kyushu Islands) in the year 2012. The volume of ballast water transferred from Japan to Europe (Atlantic, North Sea and Mediterranean coasts) was 119,762 tons. On the other hand, the volume of ballast water transferred from Japan to East Asia was 76,648,694 tons, and thus 640 times larger than the volume from Japan to Europe. Also, the volume of ballast water transferred to Japan from East Asia was 5428,768 tons, which was 45 times larger than the volume of ballast water transferred from Japan to Europe. If we take the volume of ballast water from Japan to Europe as the invasion potential of H. takanoi toward Europe, we can say that the potential of their domestic invasion to/from Japan was 1–3 orders of magnitude larger than the invasion potential toward Europe. Similarly, Kikuchi (2001) also showed that East Asia was the most frequent source of ballast water coming into Japan in the year 1997, suggesting that the estimates of Omura et al. (2014) were not a byproduct of chance. Thus we may confidently consider that shipping has indeed been active across the native range of H. takanoi, and that propagule pressure via shipping may be greater in the case across the native range compared with the case toward Europe. The second piece of evidence is related to the geographical distances to be carried for H. takanoi, which are much shorter in the case across the native Asian range compared with the case toward Europe. It is therefore expected that H. takanoi are translocated across their native range much more quickly than they are translocated toward Europe. Thus the survivorships of H. takanoi during the anthropogenic translocation would be higher in the former case than the latter case. The high individual survivorship may result in the high colonization success in the new locality. Finally, the third piece of evidence is based on the current situation: even though the propagule pressure is small and the travel time is long, artificial translocation has already enabled H. takanoi populations to successfully colonize in a remote European area, possibly multiple times (Markert et al. 2014). Thus domestic invasion of H. takanoi may have occurred more frequently compared with invasions toward Europe, given the large propagule pressure and short traveling time in the native range of H. takanoi.
However, the present results did not support this expectation due to the lack of contemporary gene flow between Japan and the area around the Yellow Sea. In other words, we may say that H. takanoi individuals from Japan cannot colonize successfully in the area around the Yellow Sea and vice versa. This result might be explained from the perspective of the dispersal–gene flow paradox (e.g., Boileau et al. 1992; De Meester et al. 2002). In short, founding populations of a species may rapidly increase their abundance in the new habitats, resulting in the blocking of successful colonization of the same species in later arrivals (see Fraser et al. 2015). This “blocking” process, hereinafter referred to as intra-specific priority effects, may occur with (e.g., monopolization hypothesis; De Meester et al. 2002) or without (e.g., persistent founder effect hypothesis; Boileau et al. 1992) the rapid local adaptation to biotic/abiotic environments there, and in either case gene flow between the dispersers and resident individuals should not become visible. By this rationale, we could argue that the intra-specific priority effects of resident H. takanoi populations may have overwhelmed the effects of artificial translocation between Japan and the area around the Yellow Sea (as shown in Fig. 5a, in contrast to Fig. 5b). As a result, genetic admixture across the East China Sea and the Tsushima Straits may not have become visible. It has been shown that H. takanoi have ecological characteristics such as early sexual maturity (ca. 10–11 months after larval settlement; Okamoto and Kurihara 1987), high fecundity (up to 60,000 eggs per brood; Okamoto and Kurihara 1987), and long reproductive period (6 months; Gothland et al. 2014) for multiple breeding (Pillay and Ono 1978). These characteristics could be strong enough to produce intra-specific priority effects in the neutral sense (Boileau et al. 1992) in the native Asian range. Alternatively but not exclusively, the fitness of H. takanoi individuals in a local population might greatly decline when they are translocated to a different local population due to maladaptation to the recipient habitat (De Meester et al. 2002).
Scheme showing the balance between artificial translocation and intra-specific priority effects in H. takanoi in their introduced and native ranges. Rectangles denote hypothetical ranges, and T0, T1, …, denote the elapsed time. Dots in the rectangles denote a group of small numbers of H. takanoi, whose genetic cluster is represented by the dot color. The genetic clusters of the native populations 1 and 2 are expressed in white and black, respectively, and “gray” dots emerge as the result of genetic admixture between the two native populations. a Within the native Asian range, geographical distances to be carried by artificial translocation are much shorter than the distances for the translocation to Europe, implying that artificial translocation would have frequently transported H. takanoi across their Asian native ranges. However, if the strength of artificial translocation is weaker than that of intra-specific priority effects of a recipient population, the artificially-transported individuals would not be able to successfully settle into the recipient population. As a result, genetic admixture would not become visible. b Alternatively, within the native range, if the strength of artificial translocation of H. takanoi is stronger than the intra-specific priority effects of a recipient population, the sign of genetic admixture might become visible at the time of this study. c In the introduced European range, the relative importance of intra-specific priority effects on the colonization success of artificially-transferred H. takanoi may have been strongly decreased, because the crabs had been transferred to an area where there had been no established H. takanoi populations. As a result, the sign of genetic admixture might become visible at the time of this study
Future directions
One of the most important findings in the present study is the genetically new, “man-made” lineages of H. takanoi in the Bay of Seine. They should be carefully monitored in the future, since genetic admixture of multiple source populations may accelerates range expansion in non-indigenous organisms (e.g., Simon-Bouhet et al. 2006; Wagner et al. 2017). In addition, further insights into the founding process of these lineages would upgrade our understanding of biological invasions. The founding events of a species in the introduced range are very different from those within the species’ native range, because in the introduced range individuals are translocated to a place where there are no or almost no established conspecific populations. Thus the relative importance of intra-specific priority effects (i.e., blocking processes) for founding populations may be strongly decreased outside of the native range. If this is the case, genetic admixture might occur smoothly during the founding process (e.g., the case in Fig. 5c), and this mechanism might be applicable to the “man-made” lineages of H. takanoi in the Bay of Seine. We could also consider that weakened intra-specific priority effects in the introduced range might also result in increased genetic variations there (but not necessarily in an additive manner), if individuals from discrete source populations arrive somewhat simultaneously. Such ideas have not been specifically examined because the role of intra-specific priority effects is rarely considered in studies of biological invasions (see Fraser et al. 2015). Thus further studies are clearly warranted.
Finally we should also mention uncertainty in the geographical distribution of H. takanoi. The present study found Hemigrapsus crabs whose morphological features did not fully match those of H. takanoi from Taiwan. Not only Hemigrapsus crabs on Taiwan but also those on the Amami Oshima Island possessed very distinct mt16S haplotypes compared with those of H. takanoi. It may be important that we did not collect H. takanoi on either island despite the fact that these islands are within the assumed geographical range of H. takanoi (see Asakura and Watanabe 2005). Although a decade has passed since the description of H. takanoi, their geographical distributions are not understood completely, and further efforts are still needed to obtain this very basic information, especially in the southern region. In such future works, the relevance of past oscillatory changes in the marine environments at the evolutionary time scale to the diversification between H. takanoi sensu stricto and closely-related Hemigrapsus crabs would also be of significant importance.
Change history
19 February 2018
It is our mistake that the Acknowledgements in the original version did not include Jean-Claude Dauvin, Jean-Philippe Pezy, Benoit Gouillieux, Guy Bachelet, Karsten Reise, and Jonas Geburzi, who kindly provided us with European H. takanoi specimens.
References
Asakura A, Watanabe S (2005) Hemigrapsus takanoi, new species, a sibling species of the common Japanese intertidal crab H. penicillatus (Decapoda: Brachyura: Grapsoidea). J Crustacean Biol 25:279–292
Blakeslee AMH, McKenzie CH, Darling JA, Byers JE, Pringle JM, Roman J (2010) A hitchhiker’s guide to the Maritimes: anthropogenic transport facilitates long-distance dispersal of an invasive marine crab to Newfoundland. Divers Distrib 16:879–891
Blakeslee AMH, Kamakura Y, Onufrey J, Makino W, Urabe J, Park S, Keogh CL, Miller AW, Minton MS, Carlton JT, Miura O (2017) Reconstructing the invasion history of the Asian shorecrab, Hemigrapsus sanguineus (De Haan 1835) in the Western Atlantic. Mar Biol 164:47
Boileau MG, Hebert PDN, Schwartz SS (1992) Non-equilibrium gene frequency divergence: persistent founder effects in natural populations. J Evol Biol 5:25–39
Carlton JT, Geller JB (1993) Ecological roulette: the global transport of nonindigenous marine organisms. Science 261:78–82
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Colwell RK (2013) EstimateS: statistical estimation of species richness and shared species from samples. Version 9 and earlier. User’s guide and application. https://purl.oclc.org/estimates
Coutts ADM, Dodgshun TJ (2007) The nature and extent of organims in vessel sea-chests: a protected mechanism for marine bioinvasions. Mar Poll Bull 54:875–886
Coutts ADM, Moore KM, Hewitt CL (2003) Ships’ sea-chests: an overlooked transfer mechanism for non-indigenous marine species? Mar Poll Bull 46:1510–1513
Cristescu ME (2015) Genetic reconstructions of invasion history. Mol Ecol 24:2212–2225
De Meester L, Gómez A, Okamura B, Schwenk K (2002) The monopolization hypothesis and the dispersal-gene flow paradox in aquatic organisms. Acta Oecol 23:121–135
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Fraser CI, Banks SC, Waters JM (2015) Priority effects can lead to underestimation of dispersal and invasion potential. Biol Invasions 17:1–8
Frey MA, Simard N, Robichaud DD, Martin JL, Therriault TW (2014) Fouling around: vessel sea-chests as a vector for the introduction and spread of aquatic invasive species. Manag Biol Invasions 5:21–30
Geburzi JC, Graumann G, Köhnk S, Brandis D (2015) First record of the Asian crab Hemigrapsus takanoi Asakura & Watanabe, 2005 (Decapoda, Brachyura, Varunidae) in the Baltic Sea. BioInvasions Rec 4:103–107 (in press)
Gollasch S (1999) The Asian decapod Hemigrapsus penicillatus (de Haan, 1835) (Grapsidae, Decapoda) introduced in European waters: status quo and future perspective. Helgol Meeresunters 52:359–366
Gothland M, Dauvin JC, Denis L, Dufossé F, Jobert S, Ovaert J, Pezy JP, Tous Rios A, Spilmont N (2014) Biological traits explain the distribution and colonization ability of the invasive shore crab Hemigrapsus takanoi. Estuar Coast Shelf Sci 142:41–49
Grizel H, Héral M (1991) Introduction into France of the Japanese oyster (Crassostrea gigas). J Cons Int Explor Mer 47:399–403
Hatakeyama S (2006) Kaki raisan. Bungei Shunjyu, Tokyo (in Japanese)
Hudson J, Viard F, Roby C, Rius M (2016) Anthoropogenic transport of species across native ranges: unpredictable genetic and evolutionary consequences. Biol Lett 12:20160620
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Katsanevakis S, Zenetos A, Belchior C, Cardoso AC (2013) Invading European seas: assessing pathways of introduction of marine aliens. Ocean Coast Manage 76:64–74
Kikuchi T (2001) Senpaku barasto mondai towa. Umito Anzen 509:2–11 (in Japanese)
Koganezawa A (1984) Matsushimagaki no konjyaku. Tohoku Suiken News 26:2–3 (in Japanese)
Lee S, Lee S-K, Rho HS, Kim W (2013) New report of the varunid crabs, Hemigrapsus takanoi and Sestrostoma toriumii (Crustacea: Decapoda: Varunidae) from Korea. Anim Syst Evol Divers 29:152–159
Makino W, Kan K, Sato M, Mukai Y, Kaiser F, Katsube T, Suzuki T, Urabe J (2015) Usefulness of the side of dark spots on the body surface as a diagnostic character distinguishing two morphologically similar Hemigrapsus species (Decapoda: Brachyura: Varunidae). Plankton Benthos Res 10:45–54
Marin IN (2013) New data on the distribution of hairly-clawed shore crabs of the genus Hemigrapsus (Decapoda: Varunidae) along the Russian mainland coast of the Sea of Japan. Rus J Mar Biol 39:301–305
Markert A, Raupach MJ, Segelken-Voigt A, Wehrmann A (2014) Molecular identification and morphological characteristics of native and invasive Asian brush-clawed crabs (Crustacea: Brachyura) from Japanese and German coasts: Hemigrapsus penicillatus (De Haan, 1835) versus Hemigrapsus takanoi Asakura & Watanabe 2005. Org Divers Ecol 14:369–382
Mingkid WM, Akiwa S, Watanabe S (2006) Morphological characteristics, pigmentation, and distribution of the sibling penicillate crabs, Hemigrapsus penicillatus (De Haan, 1835) and H. takanoi Asakura & Watanabe, 2005 (Decapoda, Brachyura, Grapsidae) in Tokyo Bay. Crustaceana 79:1107–1121
Miura O (2007) Molecular genetic approaches to elucidate the ecological and evolutionary issues associated with biological invasions. Ecol Res 22:876–883
Molnar JL, Gamboa RL, Revenga C, Spalding MD (2008) Assessing the global threat of invasive species to marine biodiversity. Front Ecol Environ 6:485–492
Niwa N, Sanagawa H, Otani M (2012) Neoeriocheir leptognathus found on Chinese commercial fishing bait known as “Isogani”. Cancer 21:53–55 (in Japanese)
Noël PY, Tardy E, d’Udekem d‘Acoz C (1997) Will the crab Hemigrapsus penicillatus invade the coasts of Europe? Comptes Rendus de l’Académie des Sciences Paris, Sciences de la Vie/Life Sciences 320:741–745
Nunes AL, Katsanevakis S, Zenetos A, Cardoso AC (2014) Gateways to alien invasions in the European seas. Aquat Invasions 9:133–144
Ohba T, Kato M, Kitazato H, Koizumi I, Omura A, Sakai T, Takayama T (1991) Paleoenvironmental changes in the Japan Sea during the last 85,000 years. Paleoceanography 6:499–518
Okamoto K, Kurihara Y (1987) Seasonal variation of population structure of Hemigrapsus penicillatus (De Haan) (Crustacea: Brachyura). Jpn J Ecol 37:81–89 (in Japanese with English abstract)
Omura T, Noma T, Kitabayashi K, Yoshida K, Saito H (2014) Current status of ballast water and aquatic organisms transferred from and to Japan. La Mer 52:13–22 (in Japanese with English abstract)
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pillay KK, Ono Y (1978) The breeding cycles of two species of grapsid crabs (Crustacea: Decapoda) from the north coast of Kyushu, Japan. Mar Biol 45:237–248
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rius M, Turon X, Bernardi G, Volckaert FAM, Viard F (2015) Marine invasion genetics: from spatio-temporal patterns to evolutionary outcomes. Biol Invasions 17:869–885
Schubart CD, Cuesta JA, Rodríguez A (2001) Molecular phylogeny of the crab genus Brachynotus (Brachyura: Varunidae) based on the 16S rRNA gene. Hydrobiologia 449:41–46
Seebens H, Gastner MT, Blasius B (2013) The risk of marine bioinvasion caused by global shipping. Ecol Lett 16:782–790
Simon-Bouhet B, Garcia-Meunier P, Viard F (2006) Multiple introductions promote range expansion of the mollusc Cyclope neritea (Nassariidae) in France: evidence from mitochondrial sequence data. Mol Ecol 15:1699–1711
Sokal RR, Rohlf FJ (1995) Biometry: the principles and practice of statistics in biological research, 3rd edn. W. H. Freeman and Company, New York
Szpiech ZA, Jakobsson M, Rosenberg NA (2008) ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504
Tada R (1994) Paleoceanographic evolution of the Japan Sea. Palaeogeogr Palaeoclimatol Palaeoecol 188:487–508
Takano M, Ikeda M, Kijima A (1997) Biochemical and morphological evidence of two sympatric forms, interpreted as sibling species, in the estuarine grapsid crab Hemigrapsus penicillatus (De Haan). Benthos Res 52:111–117
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Wagner NK, Ochocki BM, Crawford KM, Compagnoni A, Miller TEX (2017) Genetic mixture of multiple source populations accelerates invasive range expansion. J Anim Ecol 86:21–34
Wood CA, Bishop JDD, Davies CJ, Delduca EL, Hatton JC, Herbert RJH, Clark PF (2015) Hemigrapsus takanoi Asakura & Watanabe, 2005 (Crustacea: Decapoda: Brachyura: Grapsoidea): first records of the brush-clawed shore crab from Great Britain. BioInvasions Rec 4:109–113
Yamamoto K (2003) Furansu wo sukutta Nippon no kaki. Shogakkan Square, Tokyo (in Japanese)
Yamasaki I, Doi W, Mingkid WM, Yokota M, Strüssmann CA, Watanabe S (2011) Molecular-based method to distinguish the sibling species Hemigrapsus penicillatus and Hemigrapsus takanoi (Decapoda: Brachyura: Varunidae). J Crust Biol 31:577–581
Zibrowis H (1991) Ongoing modification of the Mediterranean marine fauna and flora, by the establishment of exotic species. Mésogée 51:83–107
Acknowledgements
Comments/suggestions/instructions from Susumu Chiba, Yuji Yamasaki, Ayako Suda, Takuya Kimura, the handling editor and anonymous referees significantly improved the manuscript, for which we are truly grateful. We are also grateful to Daiki Tazono, Akiyosi Shinada, Susumu Chiba, Takeshi Sonoda, Kentaro Watanabe, Kento Matsuo, Massa Nakaoka, Kenjiro Ui, Satoshi Takeda, Kyoko Kinoshita, Shin’ichiro Tsuchihashi, Takeshi Yuhara, Mitsuru Sato, Taeko Kimura, Atsushi Hirai, Tomohiro Koizumi, Koji Yamada, Tetsuya Watanabe, Jun’ya Tachikawa, Naotaka Miyajima, Tomoyuki Miura, Toru Kobari, Motohiro Shimanaga, Yoshio Kawamura, Yosuke Yamaguchi, Jiro Kawahara, Hiroyuki Doi, Yuji Tomaru, Masayuki Osawa, Masamu Fujiwara, Osamu Inamura, Mitsuhiro Fuwa, Tomoharu Kimura, Yoshihiko Machida, Ken Sakaguchi, Kotaro Kan, Masanori Sato, Shin’ichi Sato, S. Wijnhoven, J. Beermann, Maarten Boersma, and Ryuji J. Machida for helping us collect Hemigrapsus crabs. Invaluable suggestions and critical comments from AMH Blakeslee greatly improved the manuscript, for which we are grateful. The present study was supported by the Mitsui & Co., Ltd. Environmental Fund (F11-F1-020/R14-1009) and research project funds “Tohoku Ecosystem-Associated Marine Sciences (TEAMS)” from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Makino, W., Miura, O., Kaiser, F. et al. Evidence of multiple introductions and genetic admixture of the Asian brush-clawed shore crab Hemigrapsus takanoi (Decapoda: Brachyura: Varunidae) along the Northern European coast. Biol Invasions 20, 825–842 (2018). https://doi.org/10.1007/s10530-017-1604-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-017-1604-0