Abstract
Heterologous biosynthesis has been long pursued as a viable approach for high efficiency production of natural products with various industrial values. Conventional methods for heterologous biosynthesis use the mono-culture of an engineered microbe for accommodating the whole target biosynthetic pathway to produce the desired product. The emergence of modular co-culture engineering, which divides the pathway between multiple co-culture strains, presents a new perspective to conduct heterologous biosynthesis and improve the bioproduction performance of natural products. This review highlights recent advances in utilizing the modular co-culture engineering approaches to address the challenges of plant and fungal natural product biosynthesis. Potential directions for future research in this promising field are also discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant and fungal natural products have a long history of being used as a major source of nutraceutical and pharmaceutical molecules (Cragg and Newman 2013; Gupta 1994; Hoffmeister and Keller 2007; Lee 2004; Schueffler and Anke 2014). Traditionally, these biological molecules are obtained from plants or fungi through complex extraction and purification processes, which often suffer from low production yield. The relatively slow growth rate of plants and fungi as well as the difficulties associated with their metabolic engineering also largely limit the high-productivity natural product biosynthesis using these native hosts. As such, heterologous biosynthesis using a surrogate host has been developed as an alternative and viable method for generation of the plant and fungal natural products (Ahmadi and Pfeifer 2016; Becker and Wittmann 2016; Li et al. 2018a; Luo et al. 2015; Zhang et al. 2008, 2011, 2016). Specifically, a target natural product biosynthetic pathway is introduced into a selected heterologous host which is genetically and metabolically engineered to support the biosynthesis of the pathway products. This approach has been successfully utilized to make tremendous accomplishments in the past decades. Yet, it often relies on the use of a particular microbial host for accommodation of the entire complex biosynthetic pathways for targeted natural products. As such, it encounters major challenges such as imbalanced expression of pathway enzymes, impaired host cell growth due to overwhelming metabolic burden, and lack of flexibility to simultaneously satisfy the need of different heterologous and endogenous enzymes. Recent development of biosynthesis of complex natural products, especially those with highly complicated biosynthetic pathways, calls for more sophisticated methodologies for meeting the need of higher biosynthesis performance.
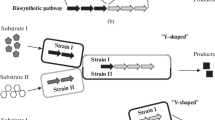
To this end, modular co-culture engineering has emerged as an alternative approach for biosynthesis of a wide range of natural products (Jones and Wang 2017; Zhang and Wang 2016). Specifically, individually engineered strains of a designed co-culture system are used to harbor different modules of the target natural product biosynthetic pathway. Owing to the versatile pathway modularization in the context of the co-culture, this methodology offers important advantages for biosynthesis including lowered metabolic stress on each strain, reduced interference between pathway enzymes, flexible balancing between pathway modules and plug-and-play fashion biosynthesis. In fact, most plant and fungal natural products have a long and complex biosynthetic pathway involving a large number of pathway enzymes with various biochemical properties, which present outstanding opportunities to leverage the power of modular co-culture engineering for improving the biosynthesis performance. A wide range of various biochemicals, including simple biofuel molecules, commodity aromatics, and complex natural products, have been successfully produced using engineered microbial co-cultures, which have been summarized in previous reviews (Jones and Wang 2017; Zhang and Wang 2016). In particular, novel engineering approaches, such as employment of multiple species co-cultures (Zhou et al. 2015), simultaneous utilization of different carbon substrates (Zhang et al. 2015), and convergent pathway engineering (Liu et al. 2018a), have been adapted to improve the capability of microbial co-cultures. The focus of this review is placed on recent achievements of plant and fungal natural product biosynthesis using the modular co-culture engineering approach between 2016 and 2018.
Co-culture biosynthesis of natural products in recent years
Pinene
Pinene is a monoterpene widely found in conifers. As this compound is considered a promising biofuel candidate, its heterologous biosynthesis using renewable feedstocks is of great research interest. Niu et al. developed an E. coli–E. coli co-culture system for pinene bioproduction using a rich medium containing sucrose, peptone and yeast extract (Niu et al. 2018). The upstream strain in the co-culture was engineered for a heterologous mevalonate pathway to produce the pathway intermediate isopentenyl diphosphate, whereas the downstream strain was dedicated to functional expression of the geranyl diphosphate synthase and pinene synthase, the last two enzymes of the pathway to convert isopentenyl diphosphate to pinene. Importantly, the designated pathway genes were integrated into the chromosome of the corresponding co-culture strains and were evolved for high copy number expression. It should be noted that, isopentenyl diphosphate, in spite of carrying the diphosphate group, was confirmed to be able to travel across the cell membrane for connecting the separate pathway modules in the context of the constructed co-culture. The optimization of the inoculation ratio between the co-culture strains resulted in the production of 64.9 mg/L pinene, which was 1.9-fold higher than the mono-culture control. The biosynthesis improvement should be attributed to the reduced metabolic burden on each strain as well as the coordinated bioconversion capabilities between the co-culture strains through the strain-to-strain ratio manipulation. Interestingly, the cells of the co-culture were also harvested, centrifuged and re-suspended in a phosphate buffer to carry out the whole-cell biocatalysis conversion, which further increased the pinene biosynthesis to 166.5 mg/L.
Monacolin J and lovastatin
Lovastatin and its precursor monacolin J are polyketide natural products found in fungi such as A. terreus and P. ostreatus. Lovastatin has been successfully developed as a commercial cholesterol-reducing drug. Liu et al. developed microbial biosynthetic systems, including Pichia pastoris-Pichia pastoris co-cultures, for de novo production of monacolin J and lovastatin using methanol as the carbon substrate (Liu et al. 2018b). To this end, the complex lovastatin biosynthesis pathway was first reconstituted in the heterologous host P. pastoris by metabolic engineering approaches. Several P. pastoris–P. pastoris co-culture strategies with different pathway splitting nodes were employed for biosynthesis improvement. The inoculation ratio between the co-culture strains was also optimized for production improvement. Notably, the variation of both the pathway splitting node and corresponding co-culture strain inoculation ratio allowed for more systematic pathway balancing than using a fixed pathway modularization pattern for biosynthesis optimization. As a result, an engineered co-culture for monacolin J biosynthesis produced 93 mg/L product, and another co-culture harboring the complete lovastatin pathway produced 24.6 mg/L lovastatin. Furthermore, the co-culture biosynthesis was scaled up using a 5 L bioreactor. The optimal inoculation ratios at this scale was investigated and found to be the same with the results of the shake flask. The highest monacolin J and lovastatin production reached 593.9 and 250.8 mg/L in the monacolin J-producing and lovastatin-producing co-cultures, respectively. Compared with the mono-culture, the biosynthesis was improved by 13.4% for monacolin J and 2.2-fold for lovastatin. Although the strain-to-strain ratio fluctuation was found during the co-culture biosynthesis, the results of the bioreactor cultivation showed the scalability and promise of the developed co-culture strategies for converting methanol to high value pharmaceuticals (Table 1).
Caffeoylmalic acid
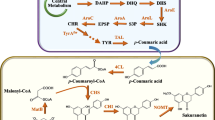
Caffeoylmalic acid, a hydroxycinnamoyl-malate ester, has been found to be an anti-oxidant and proteolytic inhibition agent and it possesses other activities beneficial for human health. This plant natural product’s biosynthesis requires the combination of caffeic acid and malate catalyzed by the hydroxycinnamoyl transferase. Li et al. reconstituted the caffeoylmalic acid pathway in E. coli by over-expression of heterologous enzymes tyrosine ammonia lyase, 4-coumarate-coenzyme A ligase, hydroxycinnamoyl transferase, enabling the de novo production on glucose (Li et al. 2018b). Moreover, an E. coli–E. coli co-culture was developed to improve the biosynthesis efficiency. In particular, the co-culture design reduced the formation of the byproduct p-coumaroylmalic acid, as the hydroxycinnamoyl transferase with promiscuous activity was put into the downstream co-culture strain and thus was segregated from the byproduct’s precursor p-coumaric acid in the upstream strain. This strategy highlighted the advantage of modular co-culture engineering for reducing the undesired interference between different pathway modules, which has also been demonstrated in previous studies (Chen et al. 2017). This effort, together with the control of the co-culture strain inoculum ratio, resulted in the production of 570.1 mg/L caffeoylmalic acid at the inoculum ratio of 6:1. Notably, the biosynthesis was three times higher than that of the mono-culture.
Resveratrol
Resveratrol is a polyphenol compound with many health-beneficial effects that makes it an attractive chemical both for academic studies and industrial purposes (Kovacic and Somanathan 2010). Co-culture biosynthesis of resveratrol precursor naringenin and other flavonoids has been reported in previous studies (Ganesan et al. 2017; Jones et al. 2017; Jones et al. 2016). To further achieve co-culture biosynthesis of resveratrol, Camacho-Zaragoza et al. adapted a co-culture system comprised of two populations of Escherichia coli strains, each with a partial and complementary section of the heterologous pathway (Camacho-Zaragoza et al. 2016). The upstream co-culture strain harbored an engineered tyrosine pathway and a heterologous tyrosine ammonia lyase for tyrosine formation and conversion to p-coumaric acid. The downstream strain was constructed to produce malonyl-CoA and provide it to the stilbene synthase STS for yielding resveratrol. The co-culture of these strains inoculated at a 1:1 ratio resulted in the production of 22.58 mg/L resveratrol from 10 g/L glycerol after 30 h. Also, p-coumaric acid was accumulated to a level lower than the mono-culture of the upstream strain, due to better conversion to resveratrol in the co-culture. Interestingly, the growth rates of both strains during co-culture cultivation were found to be similar to those of their mono-cultures, respectively.
Resveratrol glucosides
Similar co-culture design was utilized for biosynthesis of resveratrol glucosides, water-soluble derivatives of resveratrol that has been found to possess the antioxidant, estrogenic and anticancer activities (Thuan et al. 2018b). Specifically, the upstream strain of the engineered co-culture system converted exogenous p-coumaric acid to resveratrol through over-expression of 4-coumarate-coenzyme A ligase and stilbene synthase, and the downstream strains were engineered to enhance UPD-glucose formation and catalyze resveratrol glycosylation. Two glucosidated resveratrol products, polydatin and resveratroloside were successfully produced by the co-culture system with high p-coumaric acid bioconversion. Notably, the optimization of the upstream and downstream strain inoculum ratio showed that high or low ratio both increased the final polydatin production whereas the lowest production was observed at the ratio of 1:1. Nevertheless, compared to the mono-culture strategy, the co-culture improved the biosynthesis performance by 2.9-fold at the optimal inoculum ratio of 9:1, highlighting the modular co-culture engineering’s advances of straightforward balancing between individual pathway modules. Importantly, the co-culture biosynthesis was successfully scaled up using a 3 L bioreactor, leading to the production of 92.3 mg/L resveratrol glucosides.
Apigetrin
Co-culture biosynthesis has been extended for production of other flavonoids such as apigenin and its glycosylated product apigetrin, both of which possess important biological activities. Thuan and coworkers constructed an engineered E. coli–E. coli co-culture system to accommodate the apigetrin pathway (Thuan et al. 2018a). The upstream strain of the co-culture contained the pathway module for converting exogenous p-coumaric acid to apigenin through functional expression of 4-coumarate-coenzyme A ligase, chalcone synthase, chalcone flavanone isomerase, flavone synthase I. The downstream strain was specifically engineered for over-expressing UDP-glucose biosynthesis genes and its attachment to apigenin to generate apigetrin. For biosynthesis optimization, the initial inoculum ratio of the co-culture strains was varied to balance the biosynthesis capabilities of the upstream and downstream pathway modules. As a result, the apigetrin biosynthesis was improved by 2.1-fold, compared to the mono-culture strategy. In addition, the cultivation temperature was also optimized to coordinate the need of the different pathway modules. The optimized co-culture was periodically fed with exogenous p-coumaric acid and produced 16.6 mg/L apigetrin.
Salicylate 2-O-β-d-glucoside
Microbial co-cultures have also been utilized for producing other glycosylated natural products, such as a plant-based anti-inflammatory agent salicylate 2-O-β-d-glucoside. Ahmadi et al. engineered the salicylate 2-O-β-d-glucoside biosynthesis pathway in the context of E. coli–E. coli co-cultures (Ahmadi et al. 2016). In one co-culture design, the upstream strain and downstream strain were genetically modified for salicylic acid formation and glycosylation, respectively. Interestingly, it was found that the biosynthesis was not improved by this linear modularization design. To overcome this in another co-culture design, two E. coli strains, one containing the whole pathway and the other containing only the downstream glycosylation module, were co-cultivated. This strategy resulted in the production of 2.5 g/L salicylate 2-O-β-d-glucoside, which was higher than the mono-culture control. Moreover, the intermediate salicylic acid accumulation was significantly reduced in the co-culture system. This finding indicated that nonlinear pathway modularization with certain overlap between the modules may be a viable strategy for co-culture biosynthesis. Importantly, the heterologously produced salicylate 2-O-β-d-glucoside was tested and confirmed that it possessed the anti-inflammatory activity without significant mammalian cell toxicity.
Salidroside
Salidroside is a glucoside of tyrosol with demonstrated medicinal values. This natural product biosynthesis requires a convergent pathway that involves tyrosol formation and glycosylation. Liu et al. constructed an E. coli–E. coli co-culture to overcome the challenge of salidroside biosynthesis (Liu et al. 2018a). In this system, an aglycone strain was specifically engineered for tyrosol biosynthesis, and a glycoside strain was constructed to overproduce UDP-glucose and attach it to tyrosol for salidroside formation. These two strains were also genetically modified to establish the syntrophic co-culture with a phenylalanine-tyrosine cross-feeding mechanism. In order to reduce the growth competition for carbon source and improve the co-culture stability, the two strains were engineered to grow on glucose and xylose, respectively. Importantly, the glucose/xylose ratio and inoculation ratio of two strains were both adjusted for biosynthesis optimization. The optimized system was able to keep the pathway intermediate tyrosol in a low level during the co-culture cultivation. The adaption of the fed-batch bioreactor technique further improved the salidroside production to 6.03 g/L after 129 h cultivation, which demonstrated the scalability of the constructed syntrophic co-culture system. It should be noted that the modular nature of the co-culture design allowed for flexible swapping of the UDP-glycosyltransferase in the downstream strain, which led to the co-culture production of another glycoside, icariside D2.
Cadaverine
Microbial co-culture biosynthesis of cadaverine, a diamine involved in plant growth, has been reported using an E. coli–E. coli co-culture (Wang et al. 2018). In this system, two E. coli strains were used for l-lysine production and conversion to cadaverine, respectively. The two E. coli strains were also engineered to grow on glucose and glycerol, respectively, which reduced their growth competition for the carbon substrates and increased the growth balance. The co-culture cultivation conditions, including inoculation ratio, glucose/glycerol ratio, induction time, temperature, nitrogen source and concentration, C/N ratio were systematically optimized for cadaverine production. The cultivation of the engineered co-culture system in a 7.5 L fermenter led to the production of 28.5 g/L cadaverine, which was 2.1-fold higher than the mono-culture system. The biosynthesis improvement can be largely attributed to the fact that the two co-culture strains were rationally modified using metabolic engineering and bioprocess engineering tools to specifically suit the needs of the corresponding biosynthetic tasks. Importantly, the co-culture biosynthesis using the fermentor demonstrated the applicability of the microbial co-cultures at large scales.
Multi-species co-culture for cadaverine, pipecolic acid and naringenin biosynthesis
Multi-species co-cultures have also received increasing research interest, as they combine the biosynthesis powers of different species for better serving the purpose of production optimization. For example, C. glutamicum is known for robust lysine biosynthesis capability, which makes it useful for producing lysine-derived natural products. Sogbba et al. established an E. coli–C. glutamicum consortia to convert starch to pipecolic acid and cadaverine via l-lysine in one consolidated culture (Sgobba et al. 2018). Specifically, E. coli and C. glutamicum were co-cultured under the cultivation conditions that was set up to coordinate the growth between the two species. In addition, the two strains were engineered to generate a mutualistic system for improving the co-culture stability. To this end, heterologous α-amylase from S. griseus was introduced into starch-negative E. coli, allowing it to utilize starch as carbon source. The resulting glucose converted from starch by E. coli was used to feed C. glutamicum. Meanwhile, C. glutamicum provided lysine to lysine auxotrophic E. coli to facilitate its growth.
Such constructed co-culture was first employed for cadaverine production. It should be noted that, in this co-culture system, the E. coli strain was only a starch degrader, and it did not directly participate in the cadaverine biosynthesis via lysine. By deleting gene snaA encoding spermi(di)ne N-acetyltransferase and introducing lysine decarboxylase gene ldcC, C. glutamicum was engineered to enhance cadaverine bioproduction in vivo. For pipecolic acid production, gene proC encoding endogenous pyrroline 5-carboxylic acid reductase and lysDH endocing l-lysine-6-dehydrogenase were over-expressed to attract more carbon flow to pipecolic acid pathway in C. glutamicum. Based on these efforts, the production of cadaverine and pipecolic acid by the engineered co-cultures reached 6.8 mM and 3.4 mM, with the yield of 0.025 g/g (starch) and 0.012 g/g (starch), respectively.
Multi-species co-cultures have also been developed for biosynthesis of complex natural product naringenin. Zhang et al. constructed an E. coli–S. cerevisiae co-culture to combine the biosynthetic capabilities of both the prokaryotic and eukaryotic microbes for improving naringenin bioproduction efficiency (Zhang et al. 2017). Specifically, E. coli was engineered to enhance the endogenous tyrosine pathway for high level production of amino acid tyrosine, the intermediate of the naringenin pathway; S. cerevisiae was engineered to functionally express the downstream naringenin pathway enzymes, including tyrosine ammonia lyase, 4-coumarate-coenzyme A ligase, chalcone synthase, chalcone flavanone isomerase, for naringenin production. Notably, all of downstream pathway enzymes were derived from eukaryotic organisms and thus could be better expressed in S. cerevisiae.
In addition, the constituent co-culture strains were engineered to generate the commensalism for stabilization of the co-culture population composition, which had been successfully used for co-culture biosynthesis of oxygenated taxanes, tanshinone precursors and functionalized sesquiterpenes in other studies (Zhou et al. 2015). To this end, the E. coli strain was utilized to metabolize xylose and produced acetate that could be used by S. cerevisiae as the carbon source for growth. This design removed the acetate toxicity towards E. coli and also enabled S. cerevisiae to indirectly utilize xylose for growth. After the commensalism was established in the co-culture, orthogonal experiment design was carried out to optimize the cultivation conditions, including xylose concentration, inorganic salt mixtures, yeast extract and initial yeast inoculation for the co-culture biosynthesis. On top of these efforts, this study also investigated the effect of inoculum size and inoculum ratio of S. cerevisiae and E. coli. The OD600 ratio of two microorganisms was found to change dramatically after inoculation but stabilized after 48 h. Based these engineering strategies, 21.16 mg/L of naringenin was successfully produced from xylose using the optimized co-culture system.
Conclusion and future directions
In recent years, utilization of microbial co-cultures have emerged as a novel methodology for biosynthesis of various biochemicals. Due to its outstanding advantages, rationally designed co-cultures have also been increasingly used for biosynthesis of plant and fungal natural products. To date, there have been many successful studies confirming the applicability and effectiveness of this new approach for improving natural product bioproduction performance, which are summarized in this and previous reviews (Jones and Wang 2017; Zhang and Wang 2016). Therefore, further development of the modular co-culture engineering will provide a robust toolkit for overcoming the challenges of using engineered microorganisms for high-efficiency production of complex plant natural products. In the meantime, the successful application of modular co-culture engineering in plant and fungal natural product biosynthesis will generate important techniques and knowledges for wider utility of this emerging engineering approach in other biochemicals’ heterologous biosynthesis.
One of the key issues that needs to be addressed in the future is to improve the stability of the co-culture system for bioproduction enhancement. To this end, establishing mutualism or commensalism between co-culture members can be a powerful approach and is expected to receive increasing interest. In addition, selection of appropriate microbial strains or species and controlling their growth rates using metabolic engineering approaches at a similar level is also critical for improving the co-culture stability. At large scales, advanced bioprocess engineering approaches, such as fed-batch bioreactor techniques, can be adapted for coordinating the growth of different co-culture strains for biosynthesis scale-up. Another direction for future development is to more extensively adapt multi-species co-cultures for harnessing the biosynthetic capabilities of different organisms. This is of particular interest for plant and fungal natural product biosynthesis, as the use of eukaryotic organisms in the co-culture is highly beneficial for functional expression of plant and fungal natural product pathway enzymes. Successful employment of multi-species co-cultures will require manipulation of the cultivation conditions to meet the need of all involved species. It also requires in-depth understanding of species-to-species interaction in the context of co-cultures, which can be facilitated by development of sophisticated models simulating the dynamic growth and biosynthetic behaviors of the co-culture strains. Last but not least, the design of the pathway modularization plays a vital role in adapting the modular co-culture engineering approach. The proper design should choose appropriate pathway intermediates that can travel across the cell membrane of different co-culture strains for connection of the pathway modules. Moreover, the pathway modules upstream and downstream of the chosen pathway intermediate should be relatively balanced in terms of biosynthetic labor, which will greatly facilitate the biosynthetic optimization by changing the ratio between the co-culture strains carrying these pathway modules.
Moreover, it should be noted that microbial co-cultures have been developed for mimicking complex ecological systems and promoting biosynthesis of a variety of biological molecules including plant and fungal natural products, which are well summarized in recent reviews (Marmann et al. 2014). However, most of these studies focused on the activation of silenced gene expression through manipulated strain-to-strain interaction in the co-cultures, and there was not a high level of pathway modularization and bioengineering components involved in these studies. Yet, the advances in these areas offer another perspective for exploring the biosynthetic power of microbial co-cultures.
References
Ahmadi MK, Pfeifer BA (2016) Recent progress in therapeutic natural product biosynthesis using Escherichia coli. Curr Opin Biotechnol 42:7–12. https://doi.org/10.1016/j.copbio.2016.02.010
Ahmadi MK, Fang L, Moscatello N, Pfeifer BA (2016) E coli metabolic engineering for gram scale production of a plant-based anti-inflammatory agent. Metab Eng 38:382–388. https://doi.org/10.1016/j.ymben.2016.10.001
Becker J, Wittmann C (2016) Systems metabolic engineering of Escherichia coli for the heterologous production of high value molecules-a veteran at new shores. Curr Opin Biotechnol 42:178–188. https://doi.org/10.1016/j.copbio.2016.05.004
Camacho-Zaragoza JM, Hernandez-Chavez G, Moreno-Avitia F, Ramirez-Iniguez R, Martinez A, Bolivar F, Gosset G (2016) Engineering of a microbial coculture of Escherichia coli strains for the biosynthesis of resveratrol. Microb Cell Fact 15:163. https://doi.org/10.1186/s12934-016-0562-z
Chen Z, Sun X, Li Y, Yan Y, Yuan Q (2017) Metabolic engineering of Escherichia coli for microbial synthesis of monolignols. Metab Eng 39:102–109. https://doi.org/10.1016/j.ymben.2016.10.021
Cragg GM, Newman DJ (2013) Natural products: a continuing source of novel drug leads. Biochem Biophys Acta 1830:3670–3695. https://doi.org/10.1016/j.bbagen.2013.02.008
Ganesan V, Li Z, Wang X, Zhang H (2017) Heterologous biosynthesis of natural product naringenin by co-culture engineering. Synth Syst Biotechnol 2:236–242. https://doi.org/10.1016/j.synbio.2017.08.003
Gupta SS (1994) Prospects and perspectives of natural products in medicine. Indian J Pharmacol 26:1–12
Hoffmeister D, Keller NP (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep 24:393–416. https://doi.org/10.1039/b603084j
Jones JA, Wang X (2017) Use of bacterial co-cultures for the efficient production of chemicals. Curr Opin Biotechnol 53:33–38. https://doi.org/10.1016/j.copbio.2017.11.012
Jones JA et al (2016) Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab Eng 35:55–63. https://doi.org/10.1016/j.ymben.2016.01.006
Jones JA et al (2017) Complete biosynthesis of anthocyanins using E. coli polycultures. MBio. https://doi.org/10.1128/mbio.00621-17
Kovacic P, Somanathan R (2010) Multifaceted approach to resveratrol bioactivity: focus on antioxidant action, cell signaling and safety. Oxid Med Cell Long 3:86–100. https://doi.org/10.4161/oxim.3.2.3
Lee K-H (2004) Current developments in the discovery and design of new drug candidates from plant natural product leads. J Nat Prod 67:273–283. https://doi.org/10.1021/np030373o
Li S, Li Y, Smolke CD (2018a) Strategies for microbial synthesis of high-value phytochemicals. Nat Chem 10:395–404. https://doi.org/10.1038/s41557-018-0013-z
Li T, Zhou W, Bi H, Zhuang Y, Zhang T, Liu T (2018b) Production of caffeoylmalic acid from glucose in engineered Escherichia coli. Biotechnol Lett 40:1057–1065. https://doi.org/10.1007/s10529-018-2580-x
Liu X et al (2018a) Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides. Metab Eng 47:243–253. https://doi.org/10.1016/j.ymben.2018.03.016
Liu Y et al (2018b) Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol. Metab Eng 45:189–199. https://doi.org/10.1016/j.ymben.2017.12.009
Luo Y et al (2015) Engineered biosynthesis of natural products in heterologous hosts. Chem Soc Rev 44:5265–5290. https://doi.org/10.1039/c5cs00025d
Marmann A, Aly AH, Lin W, Wang B, Proksch P (2014) Co-cultivation: a powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar Drugs 12:1043–1065. https://doi.org/10.3390/md12021043
Niu FX, He X, Wu YQ, Liu JZ (2018) Enhancing production of pinene in Escherichia coli by using a combination of tolerance, evolution, and modular co-culture engineering. Front Microbiol 9:1623. https://doi.org/10.3389/fmicb.2018.01623
Schueffler A, Anke T (2014) Fungal natural products in research and development. Nat Prod Rep 31:1425–1448. https://doi.org/10.1039/c4np00060a
Sgobba E et al (2018) Synthetic Escherichia coli-Corynebacterium glutamicum consortia for l-lysine production from starch and sucrose. Bioresour Technol 260:302–310. https://doi.org/10.1016/j.biortech.2018.03.113
Thuan NH, Chaudhary AK, Van Cuong D, Cuong NX (2018a) Engineering co-culture system for production of apigetrin in Escherichia coli. J Ind Microbiol Biotechnol 45:175–185. https://doi.org/10.1007/s10295-018-2012-x
Thuan NH, Trung NT, Cuong NX, Van Cuong D, Van Quyen D, Malla S (2018b) Escherichia coli modular coculture system for resveratrol glucosides production. World J Microbiol Biotechnol 34:75. https://doi.org/10.1007/s11274-018-2458-z
Wang J et al (2018) A novel process for cadaverine bio-production using a consortium of two engineered Escherichia coli. Front Microbiol 9:1312. https://doi.org/10.3389/fmicb.2018.01312
Zhang H, Wang X (2016) Modular co-culture engineering, a new approach for metabolic engineering. Metab Eng 37:114–121. https://doi.org/10.1016/j.ymben.2016.05.007
Zhang H, Wang Y, Pfeifer BA (2008) Bacterial hosts for natural product production. Mol Pharm 5:212–225. https://doi.org/10.1021/mp7001329
Zhang H, Boghigian BA, Armando J, Pfeifer BA (2011) Methods and options for the heterologous production of complex natural products. Nat Prod Rep 28:125–151. https://doi.org/10.1039/c0np00037j
Zhang H, Pereira B, Li Z, Stephanopoulos G (2015) Engineering Escherichia coli coculture systems for the production of biochemical products. Proc Natl Acad Sci USA 112:8266–8271. https://doi.org/10.1073/pnas.1506781112
Zhang S, Wang S, Zhan J (2016) Engineered biosynthesis of medicinally important plant natural products in microorganisms. Curr Top Med Chem 16:1740–1754
Zhang W et al (2017) Production of naringenin from d-xylose with co-culture of E. coli and S. cerevisiae. Eng Life Sci 17:1021–1029. https://doi.org/10.1002/elsc.201700039
Zhou K, Qiao K, Edgar S, Stephanopoulos G (2015) Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol 33:377. https://doi.org/10.1038/nbt.3095
Acknowledgements
This work is supported by startup research funds provided by Rutgers, The State University of New Jersey. Tingting Chen is a recipient of CSC PhD fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, T., Zhou, Y., Lu, Y. et al. Advances in heterologous biosynthesis of plant and fungal natural products by modular co-culture engineering. Biotechnol Lett 41, 27–34 (2019). https://doi.org/10.1007/s10529-018-2619-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2619-z