Abstract
Objectives
N-Acetyl-d-neuraminic acid (Neu5Ac) is often synthesized from exogenous N-acetylglucosamine (GlcNAc) and excess pyruvate. We have previously constructed a recombinant Escherichia coli strain for Neu5Ac production using GlcNAc and intracellular phosphoenolpyruvate (PEP) as substrates (Zhu et al. Biotechnol Lett 38:1–9, 2016).
Results
PEP synthesis-related genes, pck and ppsA, were overexpressed within different modes to construct PEP-supply modules, and their effects on Neu5Ac production were investigated. All the PEP-supply modules enhanced Neu5Ac production. For the best module, pCDF-pck-ppsA increased Neu5Ac production to 8.6 ± 0.15 g l−1, compared with 3.6 ± 0.15 g l−1 of the original strain. Neu5Ac production was further increased to 15 ± 0.33 g l−1 in a 1 l fermenter.
Conclusions
The PEP-supply module can improve the intracellular PEP supply and enhance Neu5Ac production, which benefited industrial Neu5Ac production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sialic acid (SA) is a family of nine-carbon amino sugars, which are widespread in nature and found in both the deuterostome lineage of animals and in certain bacteria. SA usually occupies the terminal positions on macromolecules and cell membranes and thus plays vital roles in intercellular adhesion, signaling, and microbial attachment (Varki 2008). More than 50 types of SA have been reported (Inoue and Kitajima 2006) with the most ubiquitous being N-acetyl-d-neuraminic acid (Neu5Ac), which is considered valuable as medical precursor and food additive. Accordingly, an economic process for large-scale Neu5Ac production is required.
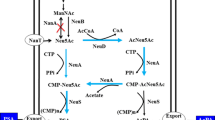
We have previously constructed a recombinant Escherichia coli strain for Neu5Ac production, in which GlcNAc 2-epimerase from Anabaena sp. CH1 (bAGE, EC 5.3.1.8) and Neu5Ac synthase from Campylobacter jejuni (cNeuB, EC 2.5.1.56, formerly EC 4.1.3.19) are coexpressed (Zhu et al. 2016). The yield of Neu5Ac was increased to 3.7 ± 0.04 g l−1 by 40 % when the GlcNAc-specific phosphotransferase system (PTS) was eliminated, indicating that the intracellular PEP supply is probably insufficient and plays a vital role in Neu5Ac production.
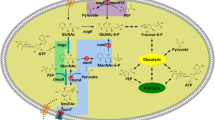
In general, the pool of intracellular PEP is stable in bacteria; PEP is synthesized under the catalysis of PEP synthase (EC: 2.7.9.2, coded by ppsA) and PEP carboxykinase (EC: 4.1.1.49, coded by pck). To improve the target product with PEP as precursor, the metabolic flux of PEP synthesis should be strengthened. We hypothesized that strengthening the PEP synthesis pathway may improve Neu5Ac biosynthesis; thus, in the current study, different PEP supply modules were constructed by overexpressing ppsA and pck to investigate their effects on Neu5Ac production (Fig. 1). Results showed that Neu5Ac production significantly improved because of the extra PEP biosynthesis pathway.
Materials and methods
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table 1. Three Duet vectors were employed as expression vectors, in which pACYCDuet-1 was used as low copy vector with a copy number of 10–12, pCDFDuet-1 and pRSFDuet-1 were used as middle copy vector and high copy vector with copy number of 20–40 and >100, respectively. E. coli strains were cultured at 37 °C with 200 rpm shaking in LB medium (yeast extract 5 g l−1, tryptone 10 g l−1, NaCl 10 g l−1, pH 7). Various combinations of ampicillin (100 mg l−1), kanamycin (50 mg l−1), streptomycin (40 mg l−1), and chloramphenicol (25 mg l−1) were added to the cultures of plasmid-bearing E. coli strains.
Analytical methods
Cell concentration was determined from the OD600 value (1 OD600 unit = 0.382 g dry cell weight l−1). Neu5Ac concentration was measured using HPLC equipped with a Bio-Rad Aminex HPX-87H column and a UV detector. Protein was determined using the method of Bradford with bovine serum albumin as standard. The expression of the recombination enzymes was verified by 12 % SDS-PAGE.
DE3 lysogenization of host Escherichia coli
Lysogenization of the host strain was carried out with λ DE3 Lysogenization Kit (Merck, Germany) with E. coli SA-04 as the original strain. First, E. coli SA-04 was cultured in lysogeny broth (LB) supplemented with 0.2 % maltose, and 10 mM MgSO4 at 37 °C to an OD600 of 0.5. Then, 1 µl pfu λ DE3, 3.3 µl pfu helper phage, and 1 µl pfu selection phage were mixed with 1 µl host cells, and the mixture was incubated at 37 °C for 30 min to allow absorption of the phages by the host cells. Afterwards, 100 µl LB medium was added to the host/phage mixture, and 10 µl of the resulting mixture was pipetted onto a LB plate and spread evenly. After incubation at 37 °C overnight, λ DE3 lysogens were verified by colony PCR.
Cloning and modification of pck and ppsA from Escherichia coli
Gene pck was obtained via PCR using E. coli MG1655 chromosomal DNA as template and pck-1-f/r as primers. The PCR product was cloned into pMD-T-simple to generate pMD-pck. Plasmid pMD-ppsA was obtained in the same manner.
The nonsense mutation of NcoI site in ppsA gene was carried out by inverse PCR using ppsA-M-f/r as primers that were composed of homologous sequences before and after the target site.
Construction of various PEP-supply modules
For the single-gene expression vector for the individual gene, both pMD-pck and pCDFDuet-1 were digested by NcoI and HindIII. Afterwards, the digested pck fragment was inserted into the corresponding site of pCDFDuet-1, generating pCDF-pck. The plasmid pCDF-ppsA was constructed in the same manner.
To construct the coexpression vector of pck and ppsA genes, the pck fragment with NdeI and KpnI tails was obtained by PCR with pck-2-f/r as primers. PCR products and plasmid pCDF-ppsA were both digested by NdeI and KpnI, followed by purification and ligation to generate the coexpression vector pCDF-ppsA-pck. Similarly, pCDF-pck-ppsA was constructed. Then, both pCDF-pck-ppsA and pRSFDuet-1 were digested by NcoI and KpnI. After purification, the products were ligated to generate the high copy coexpression vector pRSF-pck-ppsA. Similarly, pRSF-ppsA-pck, pACYC-pck-ppsA, and pACYC-ppsA-pck were constructed.
Preparation of recombinant cells and the biocatalysis process of Neu5Ac
E. coli SA-05 cells harboring both pDTrc-AB and PEP-supply module vectors were cultured in LB medium. When an OD600 of 1.2 was reached, 0.2 mM IPTG was added, and culturing was continued for 5 h to induce protein expression. To assay the enzyme activity, the induced cells were then collected and lysed by ultrasonication in an ice bath. After centrifugation, the supernatant was collected as the crude enzyme solution.
For the biocatalysis of Neu5Ac, the induced cells were collected by centrifugation (4 °C, 8000×g, 20 min), washed twice by ice-cold 0.9 % NaCl and resuspended in the reaction mixture containing 110 mM PBS (pH 7.0), 10 mM MgSO4, 0.4 M glucose, and 0.4 M GlcNAc in 50 ml (OD600nm = 30). Biocatalysis process was performed in 500 ml Erlenmeyer flasks at 37 °C with shaking at 200 rpm.
When the biocatalysis process was carried out in a 1 l fermenter (KLF 2000, Bioengineering AG, Switzerland), 15 mM PBS (pH 7.0) was used instead of 110 mM PBS, and pH was adjusted to 7 with 1 M NaOH/HCl. The fermenter was operated at 37 °C, with ventilatory capacity of 1.5 vvm and stirring at300 rpm.
Assay of enzymatic activity
AGE activity is much higher than NeuB activity (Zhu et al. 2016). NeuB activity was the probable bottleneck for Neu5Ac production. Therefore, the NeuB activity was measured.
The activity of NeuB was quantified by measuring the conversion rate of ManNAc to Neu5Ac. The reaction mixture (1 ml) consisted of Tris/HCl (0.1 M, pH 7), 10 mM MnSO4, 50 mM ManNAc, 50 mM PEP, and 20 μl crude enzyme solution. After incubation at 37 °C for 20 min, the mixture was boiled for 10 min to terminate the reaction. One unit of enzyme activity was defined as 1 nmol Neu5Ac formed per min.
Reverse transcription-quantitative PCR (RT-qPCR)
To conduct transcriptional analysis of the target genes, the total RNA was extracted from the cells after being induced with 0.2 mM IPTG for 5 h in LB. RT-qPCR was carried out as described by Kang et al. (2012). The 16 s RNA housekeeping gene was assigned as control for normalization. All primers used are listed in Supplementary Table 2.
Statistical analysis
All the experiments were performed for three times, and the statistical analysis was carried out with Student’s t test. P values of <0.05 were considered statistically significant. The specific Neu5Ac production rate was calculated by Origin software (Version 8.6, OriginLab Corp., USA).
Results and discussion
Cloning and expression of ppsA and pck in Escherichia coli
With E. coli MG1655 chromosomal DNA as template, two ORFs of 1623 and 2379 bp for pck and ppsA were amplified, respectively. The nonsense mutation by inverse PCR was introduced to mutate C (777 bp in ppsA sequence) to G to eliminate the NcoI cutting site in ppsA gene. The authenticities of these fragments were identified by dideoxy DNA sequencing (Sangon, Shanghai, China).
The Duet vectors are designed to coexpress two target proteins in E. coli, and they carry compatible replicons and antibiotic resistance markers (Ajikumar et al. 2010); certain combinations of Duet vectors and pTrc99a vectors are also compatible for coexpression. These advantages make Duet vectors ideal as the overexpression vectors for ppsA and pck even in the presence of pDTrc-AB in E. coli. The expression vectors pCDF-pck and pCDF-ppsA were constructed using the method mentioned above and verified by dideoxy DNA sequencing, respectively.
The lysogenization of host strain was verified by colony PCR with DE3-f/r as primers and was named as E. coli SA-05. When E. coli SA-05/pCDF-pck was induced, the target protein PEP carboxykinase was expressed well, whereas the target protein was not observed in the control strain E. coli SA-04/pCDF-pck lacking T7 polymerase (Fig. 2).
Construction of PEP-supply module with different expression vectors
Copy numbers of vectors and the gene order sequence of the two genes in the vectors affected gene expression. Therefore, to obtain the optimal PEP-supply module, vectors with different copy numbers were used: pACYCDuet-1 (low copy, copy number 10–12), pCDFDuet-1 (medium copy, copy number 20–40), and pRSFDuet-1 (high copy, copy number >100). For each vector, two expression vectors with different gene order sequences were constructed (Fig. 2). The expression vectors were verified by dideoxy DNA sequencing.
Single gene overexpression of pck and ppsA for the biocatalysis of Neu5Ac
For the biocatalysis of Neu5Ac, pCDF-pck and pCDF-ppsA were separately transformed into Neu5Ac producing strain E. coli SA-05/pDTrc-AB. Compared with the original strain, the PEP supply modules with the single-gene overexpression of pck and ppsA increased the yield of Neu5Ac by 61 and 96.4 %, reaching 5.8 ± 0.16 and 7.1 ± 0.24 g l−1, respectively (Fig. 2A–C).
The above figures showed that the overexpression of ppsA gene induced more promotional effects on Neu5Ac production, indicating that PEP synthase could promote the supply of intracellular PEP more directly. The overexpression of pck could also increase PEP supply; however, the substrates for PEP regeneration could be limited by the TCA cycle and the anaplerotic regulation of PEP carboxylase. Meanwhile, the glyoxylate shunt can also reduce PEP regeneration (Yang et al. 2003). As shown in Fig. 2B, C, the strains with pCDF-pck and pCDF-ppsA both showed enhanced transcription levels of corresponding genes, thereby increasing the activities of intracellular PEP carboxykinase and PEP synthase, as well as the related metabolic flux. Therefore, intracellular PEP supply and Neu5Ac production were improved. However, as PEP occupies a key branch node in the metabolites of carbohydrate with complex feedback regulation system (Tabe-Bordbar and Marashi 2013; Yang et al. 2003), when the single-gene overexpression of pck or ppsA was conducted in E. coli, the transcription level of the other gene was slightly reduced.
Co-overexpression of pck and ppsA for the biocatalysis of Neu5Ac
Effects of vector copy number
As shown in Fig. 2D–I, co-overexpression of both pck and ppsA genes eliminated the feedback inhibition mentioned above and the transcription levels of the two genes increased simultaneously. Theoretically, the increase in vector copy number can benefit the gene-transcription level. However, an excessively large copy number may increase the growth burden of host strains (Bentley et al. 2009; Ramírez et al. 2016; Song et al. 2016). Considering that low copy vectors could minimize the effects on host cells, they were used to construct engineered bacteria in some cases (Jones et al. 2000). Therefore, a vector with an appropriate copy number is essential to develop a highly efficient PEP-supply module.
The transcription level was positively correlated with the copy number of expression vectors, whereas the variation in Neu5Ac production differed (shown in Supplementary Fig. 1), the strain with medium copy vector pCDFDuet-1 observed a maximum Neu5Ac yield (8.6 ± 0.15 g l−1), compared with the 6.1–6.3 g l−1 from the strain with the high copy vector pRSFDuet-1. As shown in Fig. 3, the specific Neu5Ac production rate with pCDF-pck-ppsA and pCDF-ppsA-pck PEP-supply modules were much higher than those with other PEP-supply modules. In general, the transcription level of target genes was important but was not the sole factor. In the case of high copy vectors, the replication of vectors and the overexpression of exogenous genes consumed large amounts of metabolic intermediates and energy, which may cause low-efficiency intracellular energy metabolism (Nam et al. 2013; Wu et al. 2016). Unexpectedly, the Neu5Ac yield with low copy co-overexpression vectors was even slightly lower than that with single-gene overexpression vectors.
Therefore, the benefit of low copy vector was masked by its deficiency in expressing the target genes, whereas the high copy coexpression vector increased the transcription level of target genes but failed to enhance Neu5Ac production. The highest Neu5Ac yield was obtained with the PEP-supply module of medium copy number vector.
Effects of different gene order sequences
The Duet vectors have two T7 promoters and two MCS regions but only a single T7 terminator for the cloning and expression of two target genes. Therefore, as shown in Fig. 2D–I, the transcription level of the distal gene was higher than the proximal one, which was due to the fact that the distal gene was promoted by two T7 promoters, whereas the proximal gene was influenced only by the first T7 promoter. The highest Neu5Ac yield was achieved with the PEP-supply module of pCDF-pck-ppsA (Fig. 2F), in which ppsA gene was inserted into the distal MCS and the Neu5Ac yield was 8.4 % higher than that of pCDF-ppsA-pck.
Transcriptional analysis of bage and cneuB genes
As shown in Fig. 2, RT-qPCR results revealed that when the low copy vector pACYCDuet-1 was used, the transcription levels of cneuB and bage genes only slightly varied. Conversely, the transcription levels of cneuB and bage genes were significantly affected (negative) when the medium copy vector pCDFDuet-1 and high copy plasmid pRSFDuet-1 were used. The high copy vector may have significantly burdened the physiological activities of the host bacteria and thus negatively affected the expression of exogenous genes. However, although the transcription levels of cneuB and bage genes were reduced when medium and high copy plasmids were used, this did not reduce Neu5Ac production, which indicates that neither cNeuB activity nor bAGE activity was the bottleneck for the Neu5Ac production.
Neu5Ac production in fermenter
The above shake flask experiment data showed that pCDF-pck-ppsA was the optimum PEP-supply module with the highest efficiency. In a bench scale, Neu5Ac production by biocatalysis was carried out in a 1 l fermenter with induced E. coli SA-05/pDTrc-AB/pCDF- pck-ppsA. As shown in Fig. 4, the final yield of Neu5Ac reached 15 ± 0.33 g l−1, which was 74 % higher than that obtained in shake-flasks. Biomass slightly decreased from the beginning because of cell lysis in the new environment and then recovered until remaining stable. Intracellular NeuB activity showed no variation. With pH control, low titer of PBS (15 mM) in the fermenter setup, and better mass transfer in the fermenter, this new recombinant E. coli could produce more Neu5Ac as expected.
To date, different strategies have been used for Neu5Ac production (listed in Table 1). A fermentation strategy for producing Neu5Ac was developed with a yield of 7.85 g l−1. The Neu5Ac production strategy by biocatalysis involved an epimerization reaction to generate ManNAc from GlcNAc with AGE and an aldolization reaction to generate Neu5Ac with NeuNAc aldolase (NanA, EC 4.1.3.3). Such strategy is adopted in different catalytic processes, including whole-cell catalysis, immobilized enzymatic catalysis (Hu et al. 2010), and one-pot biosynthesis (Tao et al. 2011). Although this strategy is considered as the most economical method of industrial-scale Neu5Ac production, a significant drawback exists and could not be neglected. This drawback is that the excess pyruvate (usually fivefold) is indispensable to shift the reaction equilibrium toward Neu5Ac synthesis (Lin et al. 2013), which causes great waste of pyruvate and increases the environmental cost. The strategy adopted in this current study did not require pyruvate as substrate by using NeuB instead of NanA, and the biocatalysis of Neu5Ac was promoted by the consumption of PEP high-energy phosphate bond instead of excess pyruvate addition.
Conclusion
As a precursor of Neu5Ac synthesis, the supply of intracellular PEP is important for Neu5Ac production by biocatalysis. To construct an effective PEP-supply module, pck and ppsA genes of E. coli were single- and co-overexpressed. At the same time, expression vectors with different copy numbers were used, and the gene order sequence was also adjusted. Vector pCDF-pck-ppsA was the optimal PEP-supply module, resulting in Neu5Ac yield of 8.63 ± 0.15 g l−1 from E. coli SA-05/pDTrc-AB/pCDF-pck-ppsA, which was enhanced by 139 % as compared with that from the original strain. Neu5Ac yield was further increased to 15 ± 0.33 g l−1 in a 1 l fermenter. Consequently, our efficient PEP-supply module can improve the intracellular PEP supply and thus benefit Neu5Ac production.
References
Ajikumar PK, Xiao WH, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G (2010) Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 330:70–74
Bentley WE, Mirjalili N, Andersen DC, Davis RH, Kompala DS (2009) Plasmid-encoded protein: the principal factor in the “metabolic burden” associated with recombinant bacteria. Biotechnol Bioeng 102:1284–1297
Hu SY, Chen J, Yang ZY, Shao LJ, Bai H, Luo JL, Jiang WH, Yang YL (2010) Coupled bioconversion for preparation of N-acetyl-d-neuraminic acid using immobilized N-acetyl-d-glucosamine-2-epimerase and N-acetyl-d-neuraminic acid lyase. Appl Microbiol Biotechnol 85:1383–1391
Inoue S, Kitajima K (2006) KDN (deaminated neuraminic acid): dreamful past and exciting future of the newest member of the sialic acid family. Glycoconj J 23:277–290
Jones KL, Kim S-W, Keasling JD (2000) Low-copy plasmids can perform as well as or better than high-copy plasmids for metabolic engineering of bacteria. Metab Eng 2:328–338
Kang JH, Gu PF, Wang Y, Li YK, Yang F, Wang Q, Qi QS (2012) Engineering of an N-acetylneuraminic acid synthetic pathway in Escherichia coli. Metab Eng 14:623–629
Lin BX, Zhang ZJ, Liu WF, Dong ZY, Tao Y (2013) Enhanced production of N-acetyl-d-neuraminic acid by multi-approach whole-cell biocatalyst. Appl Microbiol Biotechnol 97:4775–4784
Lundgren BR, Boddy CN (2007) Sialic acid and N-acyl sialic acid analog production by fermentation of metabolically and genetically engineered Escherichia coli. Org Biomol Chem 5:1903–1909
Nam HK, Choi JG, Lee JH, Kim SW, Oh DK (2013) Increase in the production of beta-carotene in recombinant Escherichia coli cultured in a chemically defined medium supplemented with amino acids. Biotechnol Lett 35:265–271
Ramírez EA, Velázquez D, Lara AR (2016) Enhanced plasmid DNA production by enzyme-controlled glucose release and an engineered Escherichia coli. Biotechnol Lett 38:651–657
Song W, Bahn SY, Cha HJ, Pack SP, Choi YS (2016) Recombinant production of a shell matrix protein in Escherichia coli and its application to the biomimetic synthesis of spherulitic calcite crystals. Biotechnol Lett 38:809–816
Tabe-Bordbar S, Marashi S-A (2013) Finding elementary flux modes in metabolic networks based on flux balance analysis and flux coupling analysis: application to the analysis of Escherichia coli metabolism. Biotechnol Lett 35:2039–2044
Tao F, Zhang Y, Ma C, Xu P (2011) One-pot bio-synthesis: N-acetyl-d-neuraminic acid production by a powerful engineered whole-cell catalyst. Sci Rep 1:142
Varki A (2008) Sialic acids in human health and disease. Trends Mol Med 14:351–360
Wu G, Yan Q, Jones JA, Tang YJ, Fong SS, Koffas MAG (2016) Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol 34:652–664
Yang C, Hua Q, Baba T, Mori H, Shimizu K (2003) Analysis of Escherichia coli anaplerotic metabolism and its regulation mechanisms from the metabolic responses to altered dilution rates and phosphoenolpyruvate carboxykinase knockout. Biotechnol Bioeng 84:129–144
Zhu D, Zhan X, Wu J, Gao M, Zhao Z (2016) Efficient whole-cell biocatalyst for Neu5Ac production by manipulating synthetic, degradation and transmembrane pathways. Biotechnol Lett 38:1–9. doi:10.1007/s10529-016-2215-z
Acknowledgments
This research was supported by the National Natural Science Foundation of China No. 31171640, the Program of Introducing Talents of Discipline to Universities (111-2-06), the Fundamental Research Funds for the Central Universities (JUSRP51504, JUSRP51632A), Key Technology Research and Development Program of Wuxi (CLE01N1208), and Program for the Wuxi Bio-Agriculture Entrepreneurial Leader (130 Plan).
Supporting information
Supplementary Table 1—Strains and plasmids used in this study.
Supplementary Table 2—Primers used in this study.
Supplementary Fig. 1—Time courses of E. coli SA-05/pDTrc-AB harboring various PEP-supply modules.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, D., Wu, J., Zhan, X. et al. Phosphoenolpyruvate-supply module in Escherichia coli improves N-acetyl-d-neuraminic acid biocatalysis. Biotechnol Lett 39, 227–234 (2017). https://doi.org/10.1007/s10529-016-2235-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2235-8