Abstract
Simultaneous co-utilization of xylose and glucose is a key issue in engineering microbes for cellulosic ethanol production. We coupled xylose utilization with glucose metabolism by deletion of d-ribulose-5-phosphate 3-epimerase (RPE1) through pentose phosphate pathway flux. Simultaneous utilization of xylose and glucose then occurred in the engineered Saccharomyces cerevisiae strain with the xylose utilization pathway. Xylose consumption occurred at the beginning of glucose consumption by the engineered yeast without RPE1 in a mixed sugar fermentation. About 3.2 g xylose l−1 was utilized simultaneously with consumption of 40.2 g glucose l−1 under O2-limited conditions. In addition, an approximate ratio (~1:10) for xylose and glucose consumption was observed in the fermentation with different sugar concentration by the engineered strain without RPE1. Simultaneous utilization of xylose is realized by the coupling of glucose metabolism and xylose utilization through RPE1 deletion in xylose-utilizing S. cerevisiae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glucose and xylose are the most abundant sugars in cellulosic hydrolysates (Liu et al. 2013; Qin et al. 2013). Efficient utilization of the available glucose and xylose in the lignocellulosic hydrolysates is the important issue for economic cellulosic ethanol production.
Saccharomyces cerevisiae is the most widely used microorganism for ethanol production. However, xylose cannot be utilized in the native yeast without a heterologous xylose metabolic pathway which converts xylose to xylulose by one or two enzymes (Zha et al. 2012; Lee et al. 2014). Glucose and xylose were consumed sequentially in an engineered yeast with a xylose metabolic pathway due to the carbon catabolite repression or glucose repression (Li et al. 2010). The mechanism for glucose repression has been investigated extensively (Kim et al. 2010; Subtil and Boles 2012), but the elimination of glucose repression for simultaneous utilization of glucose and xylose in yeast is difficult due to an imperfect understanding of the mechanism (Kim et al. 2010).
Adaptation evolution, an irrational approach, has been used to pursue strains for simultaneous utilization of glucose and xylose (Shen et al. 2012; Mohagheghi et al. 2014). Strategies to avoid glucose repression were developed based on the cellobiose transporter (Saitoh et al. 2010; Ha et al. 2011; Zha et al. 2013). Hexose transporters have been analyzed for xylose and glucose specificity, and the results revealed that the moderately high-affinity permease allows xylose uptake at the same rate as that of glucose (Gonçalves et al. 2014). In Escherichia coli, simultaneous aerobic utilization of glucose and xylose was achieved by metabolic coupling with the help of a rational bilevel optimization algorithm, although the engineered E. coli exhibited a slower glucose consumption rate compared to the native strain (Gawand et al. 2013). This indicated that metabolic coupling of xylose utilization with glucose metabolism might be an alternative approach to simultaneous utilization of glucose and xylose in yeast.
The pentose phosphate pathway is not only an essential pathway for yeast survival, due to its production of important intermediate metabolites and NADPH, but it is also the key node for coupling xylose and glucose metabolism (Fig. 1). According to the metabolic network, deletion of d-ribulose-5-phosphate 3-epimerase (encoded by RPE1) would decrease the entry of carbon into the pentose phosphate pathway from glucose. Given the deletion of RPE1, the production of xylulose from xylose becomes the main source for synthesis of xylulose 5-phosphate, an essential intermediate for pentose phosphate pathway. In this study, we investigated the effects of the deletion of RPE1 on xylose and glucose fermentation, and the simultaneous utilization of glucose and xylose was observed in the engineered strain.
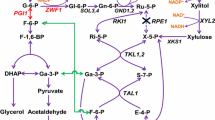
Metabolic network for glucose and xylose metabolism in recombinant xylose-utilizing Saccharomyces cerevisiae. XR xylose reductase, XDH xylitol dehydrogenase, XI xylose isomerase, XK xylulokinase, RPE ribulose-5-phosphate 3-epimerase, RKI ribose-5-phosphate ketol-isomerase, TAL transaldolase, TKL transketolase, ZWF glucose 6-phosphate dehydrogenase, SOL 6-phosphogluconolactonase, G3P glyceraldehyde 3-phosphate, S7P sedoheptulose 7-phosphate, F6P fructose 6-phosphate, E4P erythrose 4-phosphate, Gl 6-P glucono-1,5-lactone 6-phosphate, Gn 6-P gluconate 6-Phosphate, Ribu 5-P ribulose 5-phosphate
Materials and methods
Strains and media
Escherichia coli DH5α was used as a cloning host for plasmid replication, and Saccharomyces cerevisiae L2612 (MATalpha, leu2, ura3, trp1) was used as a chassis for xylose utilization (Zha et al. 2012). The yeast was grown in YNB medium (synthetic complete medium without uracil or synthetic complete medium without uracil and tryptophan) at 30 °C and 200 rpm. E. coli strains were grown in LB medium (per liter: 10 g tryptone, 5 g yeast extract, and 5 g NaCl) at 37 °C and 250 rpm. Yeast fermentation was carried out in YP medium (per liter: 10 g yeast extract, 20 g peptone) with different concentrations of xylose or/and glucose as described previously. Inocula were prepared in YNB medium.
Plasmid and strain construction
Plasmids pRS425 was used as the cloning vector. XYL1 and XYL2 genes were cloned from genome of Scheffersomyces stipitis CBS6054, and XKS1 was cloned from genome of Saccharomyces cerevisiae L2612. Primers for gene cloning were listed in Supplementary Table 1. Three expression cassettes FBA1t-TPI1p-XYL1-PGK1t, PGK1t-TPI1p-XYL2-CYC1t, CYC1t-TPI1p-XKS-TEF1t-Delta2 and Delta1-URA3- FBA1t, were constructed in pRS425 plasmid according to Lin et al. (2014). The four DNA fragments above were directly assembled and inserted into the δ-locus of L2612 (Diao et al. 2013). The strain of SyBE_Sc17002 was obtained with integration of the three xylose-utilizing genes at the δ-locus of the chromosome. The RPE1 deletion strain, SyBE_Sc17004 (RPE1::TRP1), was constructed according to one-step gene disruption strategy, and the TRP1 was cloned from plasmid pRS414 using the primers with CTAATTCCAAGAGCGAGGTAAACACACAAGAAAAAATGTCTGTTATTAATTTC and GAGAGTATAAATATAAGAAATGCCGCATATGTACAACTATTTCTTAG.
Fermentation
The seed cultures were grown in YNB medium and harvested in the late growth phase. Cells were washed twice using sterile water and inoculated into the different fermentation media, YPD (1 % (w/v) yeast extract, 2 % (w/v) peptone, and 2 % (w/v) glucose) or YPX (1 % (w/v) yeast extract, 2 % (w/v) peptone, and 2 % (w/v) xylose), starting with an OD600 = 0.1. Specific growth rates were determined in 50 ml culture in 250 ml flasks at 30 °C and 200 rpm.
For anaerobic fermentation, the seed cells were inoculated with a syringe needle to give an initial OD600 of 1 in 100 ml YPXG medium with different sugar concentrations in a 250 ml flask sealed by a rubber stopper. The cells were cultivated at 30 °C and 150 rpm. All fermentations were carried out in duplicates.
Analysis of chemicals and strain growth
Cell growth was monitored from the OD600 values. Cell dry weight was determined after drying the cell pellet in an oven at 80 °C overnight, and OD600 of 1 = 0.296 g dry cell weight l−1. Samples from fermentation were centrifuged at ~13,000×g for 5 min and the supernatant was filtered with a 0.22 µm filter. Chemicals in the sample were analyzed using HPLC with an Aminex HP-87H column (Bio-Rad, Hercules, CA, USA) at 65 °C, using 5 mM H2SO4 as eluent at 0.6 ml min−1 (Zhu et al. 2014).
Results and discussion
Xylose-utilizing yeast construction
To create a xylose-utilizing yeast, XYL1 and XYL2 genes from Scheffersomyces stipitis and XKS1 from Saccharomyces cerevisiae L2612 were assembled and inserted into the genome of L2612 at the δ locus. For the new strain, SyBE_Sc17002, an obvious lag phase for xylose utilization was observed compared with glucose utilization (Supplementary Fig. 1). The result was consistent with previous reports (Zha et al. 2014).
Deleting RPE1 to shift the strain to grow with xylose
Strain SyBE_Sc17004 was created by deleting RPE1 in SyBE_Sc17002. The strain SyBE_Sc17004 exhibited almost the same growth as SyBE_Sc17002 in the media with glucose as the sole carbon source, while no growth was observed for SyBE_Sc17004 with xylose as the sole carbon source (Fig. 2). When glucose was used as the sole carbon source, xylose 5-P is mainly generated by Rpe1p from ribulose 5-P in wild-type yeast or SyBE_Sc17002. In SyBE_Sc17004, other pathways based on Tkl1p could alternatively produce xylose 5-P from glucose due to the reversible reactions catalyzed by Tkl1p between the pentose phosphate pathway and glycolysis pathway (Fig. 1). However, when xylose was used as the sole carbon source, metabolic flux from xylose cannot enter pentose phosphate pathway or glycolysis pathway in SyBE_Sc17004, because the aid of ribose 5-P from glucose is essential for xlyose metabolism. No growth occured in the medium containing d-xylulose as the sole carbon source in the yeast strain without RPE1 (Miosga and Zimmermann 1996). According to the metabolic network for xylose and glucose (Fig. 1), metabolic shift caused by the deletion of RPE1 did not change xylose metabolism during the co-fermentation of xylose and glucose.
Triggering xylose consumption by the deletion of RPE1 at the beginning of mixed sugar fermentation
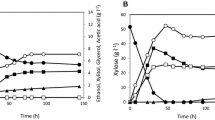
Since SyBE_Sc17004 was unable to use xylose when used as sole carbon source, a mixed sugar fermentation for SyBE_Sc17004 was carried out. As expected, xylose decreased at the beginning of glucose consumption (Fig. 3). When glucose was exhausted from 40.2 g glucose l−1, xylose concentration reduced from 10.8 g to 7.9 g xylose l−1. No xylose consumption was observed after the exhaustion of glucose, which was consistent with the growth under aerobic conditions (Fig. 2). In E. coli, the deletion of rpe and pgi resulted in the simultaneous consumption of glucose and xylose with the ratio of 1:1 (Gawand et al. 2013). No consumption of sugar was observed for the engineered E. coli strain without rpe and pgi when xylose or glucose was used as the sole carbon source (Gawand et al. 2013). In cellulosic ethanol production processes, hydrolysates from the biomass pretreated with dilute acids or steam explosion or hot water have high concentrations of glucose and low concentrations of xylose (He et al. 2014; Liu et al. 2013). Therefore, although SyBE_Sc17004 cannot consume xylose as the sole carbon source, it exhibited the potential applications for such hydrolysates with high glucose and low xylose content as mentioned above due to the potential of the simultaneous utilization of xylose and glucose for shortening the whole fermentation process.
To characterize strain SyBE_Sc17004 for sugar consumption, a series of experiments with different concentrations of mixed sugars was carried out. As is shown in Fig. 4, xylose consumption increased with the increase of glucose consumption. When the initial glucose concentration was 10 g l−1, the ratio of consumed xylose/glucose was approx 1/10. These results were consistent with the previous metabolic flux analysis, i.e., 10 % of glucose entered glycolysis through pentose phosphate pathway (Gombert et al. 2001). Xylose consumption decreased slightly based on the ratio with the increase of glucose concentration (Fig. 4), which was most likely due to the competition for transporter by high concentration of glucose (Gonçalves et al. 2014).
The stability of the ratio for xylose and glucose consumption may be due to the relative stable ratio of the flux of glycolysis and pentose phosphate pathway in the strain. When the glycolysis pathway was blocked by pgi deletion, all the flux from glucose entered pentose phosphate pathway, and xylose and glucose was consumed in the ratio of 1:1 (Gawand et al. 2013), because all the glucose entering glycolysis was through the pentose phosphate pathway and coupled the same amount of xylose due to the deletion of rpe. Only a limited proportion of glucose (10 %) was metabolized through the pentose phosphate pathway in Saccharomyces cerevisiae (Gombert et al. 2001). Such low glucose flux through the pentose phosphate pathway resulted in limited xylose consumption in the strain without RPE1. In addition, the metabolic rate of xylose in the strain SyBE_Sc17004 was still very slow. Further improvements to these yeast strains for glucose/xylose co-fermentation are needed, such as up-regulation of the non-oxidative pentose phosphate pathway flux and improvement of xylose metabolic capacity. To further improve xylose utilization, especially when xylose is the sole carbon source, some extra pathway, such as the phosphoketolase pathway (Liu et al. 2012), could be introduced into the strain without RPE1 to facilitate xylose catabolism after the depletion of glucose.
References
Diao L, Liu Y, Qian F, Yang J, Jiang Y, Yang S (2013) Construction of fast xylose-fermenting yeast based on industrial ethanol-producing diploid Saccharomyces cerevisiae by rational design and adaptive evolution. BMC Biotechnol 13:110
Gawand P, Hyland P, Ekins A, Martin VJ, Mahadevan R (2013) Novel approach to engineer strains for simultaneous sugar utilization. Metab Eng 20:63–72
Gombert AK, dos Santos MM, Christensen B, Nielsen J (2001) Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J Bacteriol 183:1441–1451
Gonçalves DL, Matsushika A, de Sales BB, Goshima T, Bon EP, Stambuk BU (2014) Xylose and xylose/glucose co-fermentation by recombinant Saccharomyces cerevisiae strains expressing individual hexose transporters. Enzyme Microb Technol 63:13–20
Ha SJ, Galazka JM, Kim SR et al (2011) Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc Natl Acad Sci USA 108:504–509
He Y, Fang Z, Zhang J, Li X, Bao J (2014) De-ashing treatment of corn stover improves the efficiencies of enzymatic hydrolysis and consequent ethanol fermentation. Bioresour Technol 169:552–558
Kim JH, Block DE, Mills DA (2010) Simultaneous consumption of pentose and hexose sugars: an optimal microbial phenotype for efficient fermentation of lignocellulosic biomass. Appl Microbiol Biotechnol 88:1077–1085
Lee SM, Jellison T, Alper HS (2014) Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol Biofuels 7:122
Li BZ, Balan V, Yuan YJ, Dale BE (2010) Process optimization to convert forage and sweet sorghum bagasse to ethanol based on ammonia fiber expansion (AFEX) pretreatment. Bioresour Technol 101:1285–1292
Lin Q, Jia B, Mitchell LA, Luo J, Yang K, Zeller KI, Zhang W, Xu Z, Stracquadanio G, Bader JS, Boeke JD, Yuan YJ (2014) RADOM, an efficient in vivo method for assembling designed DNA fragments up to 10 kb long in Saccharomyces cerevisiae. ACS Synth Biol. doi:10.1021/sb500241e
Liu L, Zhang L, Tang W, Gu Y, Hua Q, Yang S, Jiang W, Yang C (2012) Phosphoketolase pathway for xylose catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis. J Bacteriol 194:5413–5422
Liu ZH, Qin L, Jin MJ, Pang F, Li BZ, Kang Y, Dale BE, Yuan YJ (2013) Evaluation of storage methods for the conversion of corn stover biomass to sugars based on steam explosion pretreatment. Bioresour Technol 132:5–15
Miosga T, Zimmermann FK (1996) Cloning and characterization of the first two genes of the non-oxidative part of the Saccharomyces cerevisiae pentose-phosphate pathway. Curr Genet 30:404–409
Qin L, Liu ZH, Jin M, Li BZ, Yuan YJ (2013) High temperature aqueous ammonia pretreatment and post-washing enhance the high solids enzymatic hydrolysis of corn stover. Bioresour Technol 146:504–511
Saitoh S, Hasunuma T, Tanaka T, Kondo A (2010) Co-fermentation of cellobiose and xylose using beta-glucosidase displaying diploid industrial yeast strain OC-2. Appl Microbiol Biotechnol 87:1975–1982
Shen Y, Chen X, Peng B, Chen L, Hou J, Bao X (2012) An efficient xylose-fermenting recombinant Saccharomyces cerevisiae strain obtained through adaptive evolution and its global transcription profile. Appl Microbiol Biotechnol 96:1079–1091
Subtil T, Boles E (2012) Competition between pentoses and glucose during uptake and catabolism in recombinant Saccharomyces cerevisiae. Biotechnol Biofuels 5:14
Zha J, Hu ML, Shen MH, Li BZ, Wang JY, Yuan YJ (2012) Balance of XYL1 and XYL2 expression in different yeast chassis for improved xylose fermentation. Front Microbiol 3:355
Zha J, Li BZ, Shen MH, Hu ML, Song H, Yuan YJ (2013) Optimization of CDT-1 and XYL1 expression for balanced co-production of ethanol and xylitol from cellobiose and xylose by engineered Saccharomyces cerevisiae. PLoS One 8:e68317
Zha J, Shen MH, Hu ML, Song H, Yuan YJ (2014) Enhanced expression of genes involved in initial xylose metabolism and the oxidative pentose phosphate pathway in the improved xylose-utilizing Saccharomyces cerevisiae through evolutionary engineering. J Ind Microbiol Biotechnol 41:27–39
Zhu JQ, Qin L, Li BZ, Yuan YJ (2014) Simultaneous saccharification and co-fermentation of aqueous ammonia pretreated corn stover with an engineered Saccharomyces cerevisiae SyBE005. Bioresour Technol 169:9–18
Acknowledgments
This work was funded by the National Natural Science Foundation of China (21390203) and Ministry of Science and Technology of China (“973” Program: 2013CB733600).
Supporting information
Supplementary Table 1 – Primers used in the study.
Supplementary Figure 1 - Fermentation performance of SyBE_Sc17002 in medium containing 40 g glucose l−1 and 10 g xylose l-1. The initial cell density was adjusted to OD600 = 1.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shen, MH., Song, H., Li, BZ. et al. Deletion of d-ribulose-5-phosphate 3-epimerase (RPE1) induces simultaneous utilization of xylose and glucose in xylose-utilizing Saccharomyces cerevisiae . Biotechnol Lett 37, 1031–1036 (2015). https://doi.org/10.1007/s10529-014-1759-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-014-1759-z