Abstract
Biodesulphurisation was investigated by using Enterobacter sp. D4, which can selectively desulphurise and convert dibenzothiophene into 2-hydroxybiphenyl (2-HBP). The experimental values of growth, substrate consumption and product generation were obtained at 95 % confidence level of the fitted values using three models: Hinshelwood equation, Luedeking–Piret and Luedeking–Piret-like equations. The average error values between experimental values and fitted values were less than 10 %. These kinetic models describe all the experimental data with good statistical parameters. The production of 2-HBP in Enterobacter sp. was by “coupled growth”.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
SO2, which is abundant in the combustion of fossil fuels, can trigger acid rain and cause health, environment and economy problems. It is better to remove sulphur from fossil fuels as much as possible. Although most sulfur in fossil fuels can be removed easily by post-combustion methods, it is still very difficult to remove refractory organic sulfur. Biodesulphurisation (BDS) shows many advantages in the removal of refractory sulfur, such as mild experimental conditions, low costs, and little or no energy loss after treatment, etc. (Soleimani et al. 2007), compared with conventional physical and chemical processes. Dibenzothiophene (DBT) is one of the common model compounds in BDS (Bhatia & Sharma 2010) and is desulphurised through a sulphur-specific pathway called the “4S pathway” (Prayuenyong 2002).
Although Rhodococcus IGTS8, Rhodococcus erythropolis SHT87 (Fatemeh et al. 2010; Baek et al. 2002), and Gordonia alkanivorans RIPI90A (Li et al. 2008) are extensively studied examples, other microorganisms can also catalyse DBT desulphurisation. These include Microbacterium sp. NISOC-06 (Papizadeh et al. 2010), Pseudomonas putida CECT 5279 (Caro et al. 2008a), Mycobacterium sp. (Ishii et al. 2005) and Bacillus subtilis (Kirimura et al. 2001). Pantoea agglomerans D23W3, isolated and classified under Enterobacteriaceae, could also remove 26–71 % of sulphur from different petroleum oils (Bhatia & Sharma 2010).
BDS is limited by substrate and product inhibition which influence microbial growth, the reaction rate and efficiency of the BDS process (Caro et al. 2008a). Growing cells of P. putida CECT 5279 followed the Haldane kinetic model in the desulfurization of DBT (Caro et al. 2008b). More research should be conducted to determine the main limitations that decrease the yield of BDS kinetic models and also the mechanism by which these parameters affect BDS.
In our previous study, a novel bacterium, isolated from a coal mine, was identified as Enterobacter sp. (Guo et al. 2012). It could selectively remove sulphur from DBT. The present study aimed to perform desulphurisation dynamics with it. The three main rates of substrate consumption, product generation and bacterial growth were investigated. The kinetic parameters (including, µ max , K s, K p , α, β, Y x/s , m and Y p/s ) were determined and BDS kinetic models were also established.
Materials and methods
Chemicals
Dibenzothiophene (99 %), 2,6-dichloroquinone-4-chloroimide (Gibbs’ reagent) and 2-hydroxybiphenyl (2-HBP, 98 %) were purchased from J&K Scientific. Other reagents were of analytical grade and were obtained from various commercial sources.
Microorganism and medium
Enterobacter sp. D4 was grown in basal salt medium (BSM) at pH 6.8 which had the following composition (g l−1): KH2PO4 2.5, NaH2PO4·2H2O 2, NH4Cl 2, MgCl2·2H2O 0.2, glycerol 4, microelements solution 1 ml, DBT 0.1 mM (Caro et al. 2008a, b). DBT was originally dissolved in ethanol at 20 mM and added from this stock solution. The culture conditions were 100 ml medium in a 250 ml flask at 30 °C with shaking at 175 rpm for 72 h.
Analytical methods
Biomass
Cell dry weights were determined from the OD600 values. Uninoculated medium was used as a blank. DBT and 2-HBP Gibbs’ assay was conducted to detect the conversion of DBT to 2-HBP by the bacteria (Bhatia & Sharma 2010). The decrease in DBT concentration was detected by measuring A330 (Xu et al. 2002).
Theoretical analysis
Growth model of Enterobacter sp. D4
In the 4S pathway of BDS, 2-hydroxybiphenyl (2-HBP) and sulfate are directly used as sulfur sources for cell growth, and are not formed as secondary metabolites of bacterial fermentation. Consequently, in organisms using this pathway, cell growth is directly associated with substrate consumption and is inhibited by 2-HBP accumulation. Although the mechanisms of metabolite inhibition are not always clear, inhibition kinetics can be described by approximate expressions. For instance, inhibition in fermentative processes is described by the Hinshelwood model (Ma et al. 2006), in which the growth kinetics of D4 cells is governed by
where, μ max is the maximum specific growth rate (h−1); K s is the saturation constant (the concentration at which the specific growth rate is half of the maximum specific growth rate, mM) and K P is the product inhibition constant (mM).
Dynamic model of product formation by Enterobacter sp. D4
Products formed during cell growth are categorised into three groups: (1) products related to cell growth; (2) products partly related to cell growth; (3) products unrelated to cell growth. This concept was summarized by Luedeking and Piret, who proposed the Luedeking–Piret equation (Ma et al. 2006):
where α is the product synthesis constant coupled to the cell growth rate and β is the product synthesis constant coupled to the number of grown cells (h−1).
Dynamic model of substrate consumption of Enterobacter sp. D4
In the 4S pathway of BDS, the intermediate metabolite slightly accumulates, and the substrate is transformed to a metabolic end product called 2-HBP sulfate. During fermentation, substrate is primarily consumed for cell growth, metabolite accumulation and cell maintenance. Therefore, the substrate consumption rate can be represented by a Luedeking–Piret-like equation:
where Y x/s is the yield per unit substrate used during growth (mmol g−1), m is the cell maintenance coefficient (h−1), and Y p/s is the yield of the accumulated product per unit substrate.
Analytical methods
For 2-HBP analysis, the BDS culture supernatant (1 ml) was obtained and incubated in 80 μl Gibbs’ reagent (1 g Gibbs’ reagent in 1 l ethanol) at 30 °C. A change in colour from blue to purple after 30 min at room temperature indicated a positive reaction. The reaction was also monitored at A610 against a blank containing no DBT. For DBT analysis, 5 ml culture was mixed with an equal volume of ethyl acetate. The mixture was shaken and then centrifuged (15,000×g, 5 min). The supernatant fluid was assayed at A330 for the evaluation of the DBT content.
Temporal changes in dry cell weight (DCW \( C_{x} \), g l−1), 2-HBP concentration (\( C_{P} \), mM) and DBT concentration (\( C_{S} \), mM) were evaluated from samples collected at 6 h intervals. All measurements were conducted in triplicate. The derivatives at each time point were approximated by the quotient of the average difference between the values at two time points and the time interval. Data are reported as means ± standard deviations of triplicates.
To obtain the kinetic parameters, the experimental data were non-linearly fitted via the least squares method and Gauss–Newton method modified by the Levenberg–Marquart algorithm in MATLAB software. The desulfurisation kinetic models of Enterobacter sp. D4 were obtained by substituting the parameter estimates into Eqs. (1)–(3). To obtain the fitted temporal dynamics of 2-HBP production and DBT consumption, these kinetic models were then numerically solved by the fourth- and fifth-order Runge–Kutta method implemented in MATLAB.
Results
The temporal changes in 2-HBP (C p , mM) and DBT (C s , mM) concentrations were determined from experimental data of dry cell weight (C s , g l−1) listed in Table 1, as described in Methods. The results of non-linear fitting are summarised in Table 2. Substituting the estimated parameters in Table 2 into Eqs. (1)–(3), we obtained the following desulfurisation kinetic models of Enterobacter sp. D4.
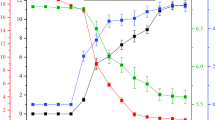
The fitted and experimental evolutions of bacterial biomass, 2-HBP concentration and DBT concentration are plotted in Fig. 1. The experimental data points are mainly observed within the 95 % confidence intervals of the fitted curves. The goodness-of-fit (R 2) values of the curves fitted to the strain growth, product formation and substrate consumption models were 0.9813, 0.9691 and 0.9691, respectively.
Comparison between fitted and actual values in the dynamic models a Bacterial concentration, b DBT consumption, and c 2-HBP production. All data are plotted as functions of time in the fermentation dynamics model. Solid round represents fitted curve; Solid triangle represents 95 % confidence low level; Solid inverted triangle represents 95 % confidence high level; Asterisk represents actual curve
Relative error analysis between the experimental and fitted values of the variables (i.e., \( C_{x} \), \( C_{P} \) and \( C_{S} \)) revealed general similarity between the experimental and fitted values (10 %), (Table 3). The Hinshelwood, Luedeking–Piret and Luedeking–Piret-like equations adequately describe the desulfurisation dynamics, and the parameters are accurately determined for each model.
The kinetic parameters of substrate consumption, Y x/s, Y p/s and m, were 62.9, 2.97 mmol g−1 and 0.00004 h−1, respectively. The strong relationship between bacterial growth and substrate consumption (Fig. 2) is consistent with the results of the kinetic model of substrate consumption (Y x/s = 62.9 mmol g−1). From the association between desulphurization product and growth (Fig. 2a), we find that the fitting parameter α (0.02) exceeds β (0.00002 h−1). Based on Gaden’s description of product formation kinetics (Gaden 1959), we classify product generation by the Enterobacter strain as Fig. 2 “coupled growth”. The temporal dynamics of the relationship between product formation and biomass in the exponential growth phase were then linearly fitted for further analysis (Fig. 2b). This result indicated an association between bacterial growth and product formation, again consistent with the kinetic model of product formation.
Comparison of the fitted lines of bacterial growth and product synthesis a product synthesis; Solid hexagon represents fitted line curve of growth D4; Solid square represents fitted line curve of DBT consumption. b Substrate consumption from 24 to 36 h; Solid hexagon represents fitted line curve of growth D4; Solid square represents fitted line curve of 2-HBP production. The experimental data of growth, substrate consumption and product generation within this period were linearly fitted
Discussion
Growth characteristics
The bacterial growth parameters were µ max = 0.16 h−1, K s = 0.5 μM and K p = 21.6 mM, indicating that the bacteria grew well in the presence of 0.1 mM DBT with minimal interference by substrate inhibition. Martin et al. (2004) studied the influence of the growth media composition on the growth and biomass production rates of Pseudomonas putida CECT 5279. Assuming the following growth kinetic model, they obtained µ max were less than 0.4 h−1 for this strain:
In a similar manner, Ma et al. (2006) obtained 0.144 h−1 and 3.88 g l−1 for the µ max and K P values. Microbial growth and BDS production were significantly inhibited by DBT reduction and 2-HBP accumulation. Caro et al. (2008a) proposed an enzymatic kinetic model with competitive inhibition in which the BDS dynamics were described by the Michaelis–Menten equation. They reported that BDS production was severely inhibited by accumulation of the 2-HBP. Hence, bacterial tolerance should be encouraged by improving the bacterial strain and biocatalyst efficiency in continuous reactors.
Desulfurisation characteristics
The kinetic parameters of substrate consumption indicated that substrate is dominantly consumed for cell growth and product synthesis. The maintenance cell coefficient (m) was very low (0.00004 h−1), demonstrating that the bacteria obtain maintenance energy from carbon sources other than DBT. This implies that the Enterobacter strain selectively oxidises sulfur atoms in DBT without cleaving the C–C bonds, thereby maintaining the caloric energy (Prayuenyong 2002).
Conclusions
We have modelled the BDS characteristics in a fermentative Enterobacter strain D4. The dynamics were well-described by kinetic models based on the Hinshelwood and Luedeking–Piret equations. These models reasonably fitted the experimental data and yielded good statistical parameters. Bacterial growth was synchronous with 2-HBP production. Kinetic parameter analysis revealed that DBT and 2-HBP inhibited bacterial growth and BDS production to different extents. Hence, the relationship between microbial growth and metabolic products could be adequately described by a growth coupling model.
References
Baek K, Kim CS, Lee HH, Shin HJ, Yang JW (2002) Microbial desulfurization of solubilized coal. Biotechnol Lett 24:401–405
Bhatia S, Sharma DK (2010) Biodesulfurization of dibenzothiophene, its alkylated derivatives and crude oil by a newly isolated strain Pantoea agglomerans D23W3. Biochem Eng J 50:104–109
Caro A, Boltes K, Letón P, García-Calvo E (2008b) Biodesulfurization of dibenzothiophene by growing cells of Pseudomonas putida CECT 5279 in biphasic media. Chemosphere 73:663–669
Fatemeh DD, Vosoughi M, Ziaee AA (2010) Biodesulfurization of dibenzothiophene by a newly isolated Rhodococcus erythropolis strain. Biores Technol 101:1102–1105
Gaden EL (1959) Fermentation process kinetics. J Biochem Microbiol Technol Eng 1(4):413–429
Guo ZG, Lu JJ, Dai DZ, Xu XL, Wei B, Sun SY (2012) Optimization of biodegradation of dibenzothiophene using response surface methodology. Trans Beijing Inst Technol 32:106–110
Ishii Y, Kozaki S, Furuya T, Kino K, Kirimura K (2005) Thermophilic biodesulfurization of various heterocyclic sulfur compounds and crude straight-run light gas oil fraction by a newly isolated strain Mycobacterium phlei Wu-0103. Curr Microbiol 50:63–70
Kirimura K, Furuya T, Nishii IY, Kino K, Usami S (2001) Biodesulfurization of dibenzothiophene and its derivatives through the selective cleavage of carbon–sulfur bonds by a moderately theromophilic bacterium Bacillus subtilis Wu-S2B. J Biosci Bioeng 91:262–266
Li GQ, Li SS, Qu SW, Liu QK, Ma T et al (2008) Improved biodesulfurization of hydrodesulfurized diesel oil using Rhodococcus erythropolis and Gordonia sp. Biotechnol Lett 30:1759–1762
Ma T, Lee K, Li GQ, Liang FL, Liu RL (2006) A kinetic model for biodesulfurization by Rhodococcus sp.DS2. J Chem Indus Eng 57:1418–1421
Martin AB, Alcon A, Santos VE, Garcia-Ochoa F (2004) Production of a biocatalyst of Pseudomonas putida CECT5279 for dibenzothiophene (DBT) biodesulfurization for different media compositions. Energy Fuels 18:851–857
Papizadeh M, Ardakani MR, Ebrahimipour G, Motamedi H (2010) Utilization of dibenzothiophene as sulfur source by Microbacterium sp. NISOC-06. World J Microbiol Biotechnol 26:1195–1200
Prayuenyong P (2002) Coal biodesulfurization processes. Songklanakarin J Sci Technol 24(3):493–507
Soleimani M, Bassi A, Margaritis A (2007) Biodesulfurization of refractory organic sulfur compounds in fossil fuels. Biotechnol Adv 25:570–596
Xu P, Ma CQ, Li FL, Tong MY, Zeng Y et al (2002) Preparation of microbial desulfurization catalysts. Chin Sci Bull 47(3):1077–1081
Acknowledgments
This study was funded by National Natural Science Foundation of China (No. 21206095) and International Cooperation Projects in Science and Technology of Xinjiang Bingtuan (Grant No. 2010YD38).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, L., Guo, Z., Lu, J. et al. Kinetic model for microbial growth and desulphurisation with Enterobacter sp.. Biotechnol Lett 37, 375–381 (2015). https://doi.org/10.1007/s10529-014-1694-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-014-1694-z