Abstract
Lung adenocarcinoma (LUAD) is a common malignancy. Many studies have shown that LUAD is resistant to gemcitabine chemotherapy, resulting in poor treatment outcomes in patients. We designed this study to reveal influences of hsa-miR-195-5p/E2F7/CEP55 axis on gemcitabine resistance and autophagy of LUAD cells. The expression data of LUAD-related mRNAs were downloaded from TCGA-LUAD database for differential expression analysis. The bioinformatics databases (hTFtarget, starBase and TargetScan) were used to predict the upstream and downstream regulatory molecules of E2F7. Then the binding relationships between E2F7 and regulatory molecules were verified by ChIP and dual-luciferase reporter assay. qRT-PCR and western blot were used to detect the mRNA and protein levels of has-miR-195-5p, E2F7, and CEP55. CCK-8 assay was used to analyze the half-maximal inhibitory concentration (IC50) and cell proliferation ability of LUAD cells after gemcitabine treatment. Apoptosis was detected by flow cytometry. Apoptosis/autophagy markers and LC3 aggregation were detected by western blot and immunofluorescence, respectively. Finally, the mouse transplantation model was constructed to verify the regulation mechanism in vivo. In LUAD cells and tissues, E2F7 and CEP55 were highly expressed, while has-miR-195-5p was relatively less expressed. The ChIP or dual-luciferase assays demonstrated the binding relationships of E2F7 to the CEP55 promoter region and has-miR-195-5p to the 3’-UTR of E2F7. Cell experiments demonstrated that overexpression of hsa-miR-195-5p stimulated LUAD cell apoptosis and inhibited autophagy and gemcitabine resistance, while further overexpression E2F7/CEP55 could reverse the impact by hsa-miR-195-5p overexpression. In vivo experiments identified that hsa-miR-195-5p/E2F7/CEP55 axis constrained the growth of LUAD tumor. Hsa-miR-195-5p promoted apoptosis, repressed proliferation, and autophagy via E2F7/CEP55 and reduced gemcitabine resistance in LUAD, indicating that hsa-miR-195-5p/E2F7/CEP55 may be a novel target for LUAD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, the relationship between apoptosis and autophagy in tumor cells has become a hot research topic. Autophagy is an intracellular self-digestion process in evolution, while apoptosis is a classical programmed death pathway, and there is a relatively complex molecular regulatory mechanism between them (Mulcahy Levy and Thorburn 2020). The activation of autophagy can utilize aging organelles, and denatured and necrotic macromolecules in tumor cells to maintain cell energy metabolism, promote the survival of tumor cells and inhibit apoptosis. Besides, autophagy can also promote apoptosis and inhibit tumor cell survival by maintaining genome stability, limiting oxidative stress, and inducing immune response (Booth et al. 2020; Yang and Klionsky 2020). Previous studies have shown that miR-138-5p inhibits autophagy of pancreatic cancer by targeting SIRT1, thus, inhibiting tumor growth (Tian et al. 2017). However, some studies have found that tumor cells can be killed by promoting autophagy. For example, SOCS5 silencing can induce autophagy activation through the PI3K/Akt/mTOR pathway, thus, inhibiting invasion and migration of liver cancer cells (Zhang et al. 2019). Thus, influences of autophagy on tumor cells are double-sided. Therefore, the exploration of this aspect can help us to understand the development of tumors and the mechanism of drug resistance.
Increasing evidence has confirmed that autophagy is important in anticancer drug therapy (Li et al. 2017). Autophagy maintains cell homeostasis by providing metabolic support for the degradation and renewal of dysfunctional organelles and long-lived proteins. Autophagy continues to occur at the normal basal metabolic level of cells. In response to starvation, hypoxia, ischemia, oxidative stress, and endoplasmic reticulum stress, autophagy can be rapidly regulated and lead to cell death (Saha et al. 2018). Tumor cells can produce resistance to anti-tumor drugs by inducing autophagy (Levy et al. 2017; Wu et al. 2015; He et al. 2015). Chen et al. (Ma et al. 2018) have found that USP9X downregulation can improve sensitivity of pancreatic cancer patients to gemcitabine by inhibiting autophagy. TNNC1 reduces the sensitivity of non-small cell lung cancer to gemcitabine chemotherapy through increasing autophagy level (Ye, et al. 2020). To conclude, autophagy exerts an imperative function in regulating chemotherapy sensitivity. Hence, we aimed to unveil the related mechanism between autophagy and tumor chemotherapy resistance.
Herein, we observed the abnormal expression states of E2F7 and hsa-miR-195-5p, and their modulatory influence on proliferation, apoptosis, autophagy, and gemcitabine resistance of LUAD cells. We also explored the relevant downstream regulatory mechanisms that may be affected by E2F7 and found that E2F7 could activate CEP55 to inhibit apoptosis of LUAD cells, stimulate autophagy, and reduce gemcitabine sensitivity. These findings contributed to a bolstered understanding of mechanisms of disease development and gemcitabine resistance and suggested that hsa-miR-195-5p/E2F7/CEP55 axis may be potential therapeutic target for LUAD.
Materials and Methods
Bioinformatics Analyses
LUAD-related mRNA expression data were downloaded from TCGA-LUAD database (normal: 59, tumor: 535) and subjected to differential expression analysis using the “edgeR” package (|logFC|> 2, FDR < 0.05) to acquire differentially expressed mRNAs (DEmRNAs). Target gene E2F7 was identified by literature citation. HTFtarget database was used to predict the potential target mRNA downstream of E2F7 in LUAD. StarBase and TargetScan were used to predict the regulatory genes upstream of E2F7. Pearson correlation analysis was used to investigate the correlation between E2F7 and the expression levels of regulatory molecules. JASPAR was employed to predict the binding sites between E2F7 and regulatory molecules.
Cell culture and Cell Transfection
Human LUAD cell lines H1299 (BNCC-100859) and A549 (BNCC-100215), HCC827 (BNCC-353294) and human bronchial epithelial cells BEAS-2B (BNCC-339275) bought from BeNa Culture Collection (BNCC, China) were placed in RPMI-1640 plus 10% fetal bovine serum (FBS). Cells were maintained under conditions of 37 ℃ and 5% CO2.
Lipofectamine 2000 Kit (GenePharmam Suzhou, China) was utilized to perform cell transfection of LUAD cells with si-E2F7, oe-E2F7, si-CEP55, oe-CEP55, hsa-miR-195-5p mimic, hsa-miR-195-5p agomir, and corresponding negative controls (GenePharmam Suzhou, China).
qRT-PCR
Total RNA extraction was performed by Trizol method (Invitrogen, USA). cDNA was then obtained and subjected to qRT-PCR on SYBR Green Master (Roche, Switzerland). β-actin was taken as an endogenous reference for E2F7 and CEP55, and U6 for hsa-miR-195-5p. Data were analyzed by 2−ΔΔCt method. Specific primer sequences are presented in Table 1.
Western Blot
Detailed steps of western blot were performed according to previous description (Chen, et al. 2021). Primary antibodies, including rabbit anti-human LC3I/II (LC3B, ab192890, 1:2000), β-actin (ab8227, 1:2000), CEP55 (ab170414, 1:5000), Bcl-2 (ab32124, 1:1000), Bax (ab32503, 1:2000), Cleaved caspase-3 (ab32042, 1:500), and p62 (ab91526, 1:2000), were purchased from Abcam (UK). Primary antibody rabbit anti-human E2F7 (DF2444, 1:2000) was accessed from Affinity (USA). Secondary antibody goat anti-rabbit IgG (ab205718, 1:50000) was purchased from Abcam (UK).
Chromatin Immunoprecipitation (ChIP)
ChIP assay was conducted as per steps in the previous article (Zhao et al. 2020). According to the kit instructions, ChIP assay was performed using IP grade anti-E2F7 antibodies (ab245655, 1: 1000), Rabbit IgG (ab172730,1:1000), and corresponding Simple ChIP enzymatic chromatin IP kit (CST, USA). qPCR was used to evaluate the purified DNA. Primers are listed in Table 2.
Dual-Luciferase Assay
Luciferase reporter vectors of pGL3-CEP55-WT and pGL3-CEP55-MUT (Promega, USA) were constructed. A pRL reference plasmid was used as a control. Then, the LUAD cell line HCC827 (2 × 105 cells/well) was plated into 96-well plates. Next, cells were co-transfected with luciferase reporter plasmid, Renilla luciferase reporter plasmid and oe-NC or oe-E2F7. 48 h later, the luciferase activity was measured using a dual-luciferase reporting system (Promega, USA). This assay was performed in triplicate.
CCK-8 Assay
CCK-8 (Beyotime Institute Biotech, China) was conducted for assessing proliferative property of LUAD cells. Transfected cells from each group were collected and prepared into cell suspensions. 100 μL cell suspensions were then plated into 96-well plates (2 × 103 cells/well), and then the plates were precultured with standard conditions. At 0, 24, 48, 72, and 96 h, CCK-8 solution (10 μL) was slowly added to the wells. Cells containing CCK-8 solution were incubated for 2 h. At the end of the incubation, absorbance value at 450 nm was assessed with a microplate reader (Sunnyvale, USA) (Zhao et al. 2021).
CCK-8 (Beyotime Institute Biotech, China) was employed to assay sensitivity of LUAD cells to gemcitabine (An et al. 2018). The IC50 value was determined as per instructions. LUAD cells (5 × 103 cells/well) were inoculated into 96-well plates for 24 h incubation. Absorbance value at 450 nm was measured with a microplate reader (Sunnyvale, USA). Gemcitabine was purchased from Merck Life Sciences (China). After treatment with Gemcitabine (0, 0.001, 0.01, 0.1, 1, 5, and 10 μg/mL) for 24 h and 48 h, respectively, IC50 value was detected.
Flow Cytometry
Apoptosis of LUAD cells was assayed via flow cytometry (BD Biosciences, USA). This assay was conducted according to the kit instructions (Invitrogen, USA). Double Annexin V/PI staining was adopted and flow cytometry was performed.
Immunofluorescence
The immunofluorescence test was performed by referring to the method described previously (Wang et al. 2018). The LC3 antibody used was the same as western blot.
Mouse Transplantation Model
20 BALB/c nude mice (4 weeks; 18–25 g) with specific pathogen-free grade were provided by SLAC Laboratory Animal Co. Ltd. (China) and then assigned randomly to four groups (five mice/group). Cells transfected with NC-agomir + oe-NC, hsa-miR-195-5p agomir + oe-NC, hsa-miR-195-5p agomir + oe-E2F7, and hsa-miR-195-5p agomir + oe-CEP55 (2 × 106 cells/mice) were injected into mice subcutaneously, and growth of tumors was recorded every 7 days. 4 weeks later, mice were euthanized and tumor tissues were removed. Volume (volume = L × W2/2) and weight of tumors were measured for subsequent experiments. The study was approved by the Ethics Committee.
Statistical Analysis
All data were expressed as Mean ± SD. In this study, difference significance among multiple groups was assessed by one-way analysis of variance (ANOVA) and that between two groups was tested by Student’s t test. Each experiment was independently repeated three times. P < 0.05 indicated significant difference.
Results
E2F7 is Highly Expressed in LUAD Tissues and Cells
LUAD-associated mRNA expression data from TCGA-LUAD database were subjected to differential analysis, with the result disclosing that E2F7 was evidently increased in LUAD tissues compared with normal tissues (Fig. 1A). The results of survival analysis illustrated that early survival rate of patients with relatively poor E2F7 expression was markedly higher than those with high E2F7 expression (Fig. 1B). Combined with previous studies, we believed that E2F7 could facilitate the malignant progression of LUAD (Wang et al. 2021; Liang et al. 2018). To verify this result, qRT-PCR and western blot assays analyzed E2F7 level in A549, H1299, HCC827, and BEAS-2B cells. It was observed that E2F7 expression in LUAD cells was noticeably up-regulated (Fig. 1C–D). The expression of E2F7 was relatively high in H1299 and HCC827 cells, so these two cell lines were selected for subsequent assays. These results suggested a significantly high expression of E2F7 in LUAD.
E2F7 is highly expressed in LUAD tissues and cells. A Expression of E2F7 in normal tissues and LUAD tissues; B The survival curve of LUAD patients based on median E2F7 expression in LUAD tissues; C–D: The mRNA and protein expression of E2F7 in LUAD cell lines (A549, H1299, HCC827) and human bronchial epithelial cells (BEAS-2B). ***, **** meant P < 0.001, 0.0001, respectively (one-way ANOVA)
Effects of E2F7 on Proliferation, Apoptosis, Autophagy, and Sensitivity to Gemcitabine of LUAD Cells
As the role of E2F7 in LUAD progression was underexplored, si-NC and si-E2F7 were transfected into H1299 and HCC827 cells, with transfection efficiency being detected via qRT-PCR and western blot. Significantly lower E2F7 expression was presented in si-E2F7 treatment group than in control (Fig. 2A–B). Later, we tested the effect of E2F7 on LUAD cell proliferation through CCK-8, which presented suppression of LUAD cell proliferation by knockdown of E2F7 (Fig. 2C). Subsequently, flow cytometry results showed that silencing E2F7 promoted apoptosis of LUAD cells (Fig. 2D). Meanwhile, we detected the expression of apoptosis-associated proteins using western blot. Silencing E2F7 reduced Bcl-2 protein expression but increased Bax and Cleaved caspase-3 protein expression in LUAD cells (Fig. 2E). Next, impact of E2F7 on autophagy of LUAD cells was unveiled. LC3 aggregation was assayed through immunofluorescence assay. It was markedly reduced in si-E2F7 group compared with control group (Fig. 2F). Meanwhile, we measured autophagy-related protein levels in LUAD cells. LC3 II/LC3 I protein expression was remarkably lowered and that of p62 protein was evidently elevated in LUAD cells in si-E2F7 group (Fig. 2G). Finally, to study influence of E2F7 on gemcitabine chemotherapy sensitivity, we measured the IC50 value of LUAD cells to gemcitabine in each group by using CCK-8 assay. We found a significantly decreased IC50 value of LUAD cells to gemcitabine in si-E2F7 group compared with the si-NC group (Fig. 2H–I), suggesting that silencing E2F7 could improve the sensitivity of LUAD cells to gemcitabine. In conclusion, E2F7 stimulated proliferation and autophagy of LUAD cells, constrained apoptosis of LUAD cells, and reduced the sensitivity of LUAD cells to gemcitabine.
Effects of E2F7 on apoptosis, autophagy, and gemcitabine chemotherapy sensitivity of LUAD cells. A, B E2F7 mRNA and protein expression in LUAD cells (H1299 and HCC827) in each group; C The proliferation ability of LUAD cells; D The apoptosis level of LUAD cells in each group; E The expression of apoptosis-related proteins in LUAD cells; F Immunofluorescence assay on the expression of E2F7 and LC3 protein in LUAD cells in each group; G The expression of autophagy-related proteins in LUAD cells; H, I The IC50 value of LUAD cells in different groups treated with gemcitabine (0, 0.001, 0.01, 0.1, 1, 5, and 10 μg/mL) for 24 h and 48 h, respectively. **, ***, **** meant P < 0.01, 0.001, 0.0001, respectively (Student's t test)
CEP55 is a Downstream Regulator of E2F7
To understand mechanism of E2F7 modulating LUAD progression, the target mRNAs of transcription factor E2F7 in LUAD were predicted by hTFtarget database. From the intersection of 145 potential target genes and 1,969 up-regulated genes, 8 DEmRNAs were identified as the potential target genes (Fig. 3A). The results of survival analysis illustrated a substantially higher survival rate of patients with low CEP55 level than those with high CEP55 level (Fig. 3B), indicating that high CEP55 level was closely implicated in unfavorable prognoses of LUAD patients. Next, Pearson correlation analysis showed that CEP55 level was significantly and positively correlated with E2F7 (Fig. 3C). Subsequently, binding sites of E2F7 to TSS region of CEP55 promoter were searched by JASPAR database. The results showed that there were two potential binding sites at the first 2000 bp of CEP55 promoter (Fig. 3D), and CEP55 was evidently overexpressed in LUAD tissues (Fig. 3E). Then, qRT-PCR was used to analyze CEP55 mRNA level in LUAD and bronchial epithelial cells, the result of which exhibited that CEP55 was remarkably increased in LUAD cells (Fig. 3F). As E2F7 had a significant effect on the apoptotic phenotype of HCC827 cell line, it was selected for further analyses. Then, ChIP result validated the binding relationship of E2F7 with CEP55 (Fig. 3G). Dual-luciferase assay verified the targeting between E2F7 and CEP55, indicating that E2F7 overexpression increased luciferase activity of wild-type CEP55 but did not alter the luciferase activity of mutant CEP55 (Fig. 3H). The expression of CEP55 after E2F7 knockdown was inhibited as detected via qRT-PCR (Fig. 3I). Hence, E2F7 could directly facilitate the transcription of CEP55.
CEP55 is a downstream regulatory molecule of E2F7. A Venn diagram of target genes of E2F7 predicted by bioinformatics analysis and differentially up-regulated genes; B The survival curve of LUAD patients based on median CEP55 expression in LUAD tissues; C Pearson correlation analysis of E2F7 and CEP55; D The binding sites of E2F7 to the TSS region of CEP55 promoter was validated by JASPAR database; E CEP55 expression in normal tissues and LUAD tissues; F The mRNA expression of CEP55 in human bronchial epithelial cells (BEAS-2B) and LUAD cell lines (A549, H1299, HCC827); G, H: CHIP and dual-luciferase assays that verifies the binding relationship between E2F7 and CEP55 promoter; I The expression level of CEP55 when E2F7 was knocked down. ***, **** meant P < 0.001, 0.0001, respectively (one-way ANOVA)
E2F7 Affects Proliferation, Apoptosis, Autophagy, and Gemcitabine Chemotherapy Resistance of LUAD Cells via CEP55
To unveil function of E2F7/CEP55 axis in LUAD progression, we set up the following experimental groups: si-NC+oe-NC, si-CEP55+oe-NC, si-NC+oe-E2F7, and si-CEP55+oe-E2F7. First, the transfection efficiency of HCC827 cells in each treatment group was determined through qRT-PCR and western blot experiments. Compared with control group, CEP55 expression was decreased in si-CEP55+oe-NC group and substantially elevated in si-NC+oe-E2F7 group, and that in si-CEP55+oe-E2F7 group was recovered (Fig. 4A–B). CCK-8 assay for assessment of proliferative property revealed the result that as compared to control, cell proliferation property of si-CEP55+oe-NC group was significantly reduced, and that of si-NC+oe-E2F7 group was significantly increased. The cell proliferation ability of si-CEP55+oe-E2F7 group was restored to the level of si-NC+oe-NC group (Fig. 4C). Then, flow cytometry for apoptosis analysis depicted the finding that apoptosis rate of LUAD cells reduced upon E2F7 overexpression and increased upon CEP55 silencing and that simultaneous CEP55 silencing and E2F7 overexpressing could restore the apoptosis level of LUAD cells (Fig. 4D). Next, apoptosis-related proteins were further detected. It was shown that CEP55 silencing remarkably elevated Cleaved caspase-3 and Bax protein levels but decreased that of Bcl-2 protein in LUAD cells and that simultaneous CEP55 silencing and E2F7 overexpressing could restore apoptosis-related protein levels in LUAD cells (Fig. 4E). Then, LC3 aggregation was assayed via immunofluorescence assay. In comparison with the control, CEP55 silencing significantly reduced LC3 aggregation in LUAD cells, but forced expression of E2F7 rescued the effect of CEP55 silencing on LC3 aggregation of LUAD cells (Fig. 4F). Meanwhile, autophagy-related protein levels in LUAD cells were assayed. Compared with si-NC + oe-NC group, CEP55 silencing prominently decreased LC3 II/LC3 I protein level and substantially elevated P62 protein level. However, E2F7 overexpression had the opposite effects. Overexpression of E2F7 reversed impact of CEP55 silencing on autophagy in LUAD cells (Fig. 4G). Finally, we investigated the effects of E2F7/CEP55 axis on gemcitabine resistance in LUAD cells. As compared to control, E2F7 overexpression and CEP55 silencing resulted in a notable increase and decrease in the IC50 value of LUAD cells to gemcitabine, respectively. However, simultaneous operation of CEP55 silencing and the E2F7 overexpression restored the IC50 value of LUAD cells response to gemcitabine (Fig. 4H–I). Hence, CEP55 silencing enhanced sensitivity of LUAD cells to gemcitabine, and further overexpression of E2F7 overturned influence of CEP55 silencing on sensitivity of LUAD cells to gemcitabine. In order to check whether overexpression of CEP55 can abrogate the phenotypes of E2F7 in LUAD cells, we constructed si-NC+oe-NC, si-E2F7+oe-NC, si-NC+oe-CEP55, and si-E2F7+oe-CEP55 transfection groups for verification. Overexpression of CEP55 did not change E2F7 level but could restore the changes of proliferation, apoptosis, autophagy, and gemcitabine sensitivity of LUAD cells induced by knockdown of E2F7, which further indicated that E2F7 regulated the malignant progression of LUAD by targeting CEP55 (Supplementary Fig. 1). In conclusion, by activating CEP55, E2F7 inhibited LUAD cell apoptosis, promoted LUAD cell proliferation and autophagy, and increased the drug resistance of LUAD cells to gemcitabine.
E2F7 affects apoptosis, autophagy, and gemcitabine resistance of LUAD cells via CEP55.A, B The mRNA and protein expression of CEP55 of HCC827 cells in each group; C The proliferation ability of LUAD cells; D The apoptosis of HCC827 cells in each group; E The expression of apoptosis-related proteins in LUAD cells; F Immunofluorescence assay of E2F7 and LC3 protein expression in HCC827 cells in each group; G The expression of autophagy-related proteins in HCC827 cells; H, I The cell viability and IC50 values respond to 24 h and 48 h of gemcitabine treatment (0, 0.001, 0.01, 0.1, 1, 5, 10 μg/mL), respectively. *, **, ***, **** meant P < 0.05, 0.01, 0.001, 0.0001, respectively (one-way ANOVA)
Hsa-miR-195-5p is an Upstream Regulatory Gene of E2F7
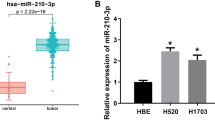
Regulatory mechanism of E2F7 in LUAD cells was to be revealed, therefore, we used starBase and TargetScan to predict the regulatory genes upstream of E2F7. The results were intersected with 16 down-regulated miRNAs, and 5 differential miRNAs were identified (Fig. 5A). Pearson correlation analysis between E2F7 and the predicted five target genes revealed that hsa-miR-195-5p was most significantly associated with E2F7, and correlation was negative (Fig. 5B). Therefore, miR-195-5p was deemed as target gene, and TCGA database analysis illustrated that it was markedly under-expressed in LUAD tissues (Fig. 5C). Subsequently, binding site of E2F7 to hsa-miR-195-5p was obtained through JASPAR database (Fig. 5D), and the targeting relationship between the two was further verified by dual-luciferase experiment. When hsa-miR-195-5p was overexpressed, dual-luciferase activity of wild-type E2F7 was substantially reduced, but dual-luciferase activity of mutant E2F7 did not change significantly (Fig. 5E), suggesting that hsa-miR-195-5p could target and bind E2F7. Then, qRT-PCR assayed mRNA level of hsa-miR-195-5p in BEAS-2B, A549, H1299, and HCC827, with the experimental results showing that hsa-miR-195-5p was notably decreased in LUAD cells (Fig. 5F). Finally, we also tested regulatory relationship between hsa-miR-195-5p and E2F7, with the result presenting that E2F7 level was noticeably reduced with overexpressed hsa-miR-195-5p (Fig. 5G). Based on the findings, we concluded that hsa-miR-195-5p was an upstream regulatory gene of E2F7 and negatively modulated E2F7 level.
Hsa-miR-195-5p is an upstream regulatory gene of E2F7. A Intersection of predicted miRNAs targeting E2F7 and differentially down-regulated miRNAs; B Pearson correlation analysis results of E2F7 and the predicted five miRNAs, and Pearson correlation analysis results of hsa-miR-195-5p and E2F7; C The expression level of hsa-miR-195-5p in LUAD tissues analyzed by TCGA database; D The binding site of E2F7 to hsa-miR-195-5p; E Dual-luciferase assay was used to detect the targeting relationship between E2F7 and hsa-miR-195-5p; F The expression of hsa-miR-195-5p in different cells; G The mRNA expression of E2F7 in cells after overexpression of hsa-miR-195-5p. **, ***, **** meant P < 0.01, 0.001, 0.0001, respectively (one-way ANOVA)
Hsa-miR-195-5p/E2F7/CEP55 Axis can Promote LUAD Cell Apoptosis, Inhibit Proliferation and Autophagy, and Reduce the Resistance of LUAD Cells to Gemcitabine
To analyze impact of hsa-miR-195-5p/E2F7/CEP55 axis on functions of LUAD cells, the following experimental groups were set: NC mimic+oe-NC, hsa-miR-195-5p mimic+oe-NC, hsa-miR-195-5p mimic+oe-E2F7, and hsa-miR-195-5p mimic+oe-CEP55. Firstly, CEP55 mRNA and protein levels in HCC827 cell treatment groups were determined via qRT-PCR and Western blot to evaluate transfection efficiency. Compared with the control group (NC mimic+oe-NC), CEP55 expression was decreased in hsa-miR-195-5p mimic+oe-NC group but was notably increased by E2F7 overexpression. The expression of CEP55 in hsa-miR-195-5p mimic+oe-CEP55 group was restored to the level of NC mimic+oe-NC group (Fig. 6A–B). CCK-8 for determination of proliferative property illustrated that compared with control, proliferation ability of hsa-miR-195-5p mimic+oe-NC group was remarkably reduced, but it was substantially increased with forced expression of E2F7/CEP55. Cell proliferation ability of hsa-miR-195-5p mimic+oe-E2F7 group and hsa-miR-195-5p mimic+oe-CEP55 group was restored to the level of NC mimic+oe-NC group (Fig. 6C). Subsequently, cell apoptosis was analyzed via flow cytometry. The apoptosis rate of cells in hsa-miR-195-5p mimic+oe-NC group was dramatically elevated, and further overexpression of E2F7/CEP55 could noticeably decrease apoptosis rate (Fig. 6D). Apoptosis-related protein detection results showed that in LUAD cells, cleaved caspase-3 and Bax protein levels were prominently up-regulated in hsa-miR-195-5p mimic+oe-NC group, but Bcl-2 protein level was in the opposite. Further overexpression of E2F7/CEP55 down-regulated Cleaved caspase-3 and Bax protein expression, and up-regulated Bcl-2 protein expression. Apoptosis-related protein levels in hsa-miR-195-5p mimic+oe-E2F7 group and hsa-miR-195-5p mimic+oe-CEP55 group were recovered (Fig. 6E). Then, the aggregation of LC3 in different treatment groups was assayed via immunofluorescence assay. LC3 aggregation in LUAD cells in hsa-miR-195-5p mimic+oe-NC group was remarkably decreased in comparison to control, and further overexpression of E2F7/CEP55 significantly increased LC3 aggregation. Intracellular LC3 aggregation was restored to the level of NC mimic+oe-NC group (Fig. 6F). Meanwhile, autophagy-related protein levels in LUAD cells were assayed. Compared with NC mimic + oe-NC group, LC3 II/LC3 I protein level was notably decreased and the protein level of P62 was substantially elevated in hsa-miR-195-5p mimic+oe-NC group. Further overexpression of E2F7/CEP55 significantly increased the intracellular protein level of LC3 II/LC3 I and significantly decreased that of P62 (Fig. 6G). Finally, we investigated the impact of hsa-miR-195-5p/E2F7/CEP55 axis on gemcitabine resistance in LUAD cells. As in comparison to control group, forced expression of hsa-miR-195-5p significantly reduced IC50 value of LUAD cells to gemcitabine, and further overexpression of E2F7/CEP55 attenuated the drug sensitivity of LUAD cells to gemcitabine, resulting in a substantial increase in IC50 value of gemcitabine in LUAD cells (Fig. 6H–I). In conclusion, hsa-miR-195-5p promoted LUAD cell apoptosis, inhibited proliferation and autophagy, and attenuated gemcitabine resistance via regulating E2F7/CEP55.
Hsa-miR-195-5p/E2F7/CEP55 axis can promote apoptosis of LUAD cells, inhibit autophagy, and reduce gemcitabine resistance in LUAD. A, B The transfection efficiency was evaluated by qRT-PCR and Western blot; C The proliferation ability of LUAD cells in each transfection group; D The apoptosis of each transfected group; E The expression of apoptosis-related proteins in each transfected group; F The aggregation of LC3 was detected by immunofluorescence assay; G The expression level of autophagy-related proteins in the transfected cells; H, I: The cell viability and IC50 values respond to 24 h and 48 h of gemcitabine treatment (0, 0.001, 0.01, 0.1, 1, 5, 10 μg/mL), respectively. **, ***, **** meant P < 0.01, 0.001, 0.0001, respectively (one-way ANOVA)
Hsa-miR-195-5p/E2F7/CEP55 Axis can Inhibit the Growth of LUAD Tumors
We have previously found that hsa-miR-195-5p/E2F7/CEP55 can inhibit malignant progression of LUAD cells through in vitro experiments. Here, we verified influence of hsa-miR-195-5p/E2F7/CEP55 axis on LUAD tumor growth in mice through animal experiments. First, we randomly divided the mice into four groups: NC-agomir+oe-NC, hsa-miR-195-5p agomir+oe-NC, hsa-miR-195-5p agomir+oe-E2F7, and hsa-miR-195-5p agomir+oe-CEP55. The cells after different treatments were subcutaneously injected into mice according to groups. Tumor growth rate and weight of hsa-miR-195-5p agomir group were notably inhibited. Compared with hsa-miR-195-5p agomir+oe-NC, the tumor growth trend and tumor weight of hsa-miR-195-5p agomir+oe-E2F7 group and hsa-miR-195-5p agomir+oe-CEP55 group were noticeably increased, which was similar to the NC-agomir+oe-NC group (Fig. 7A–C). qRT-PCR and Western blot were used to assess expression of CEP55 mRNA and protein in mice. The expression of CEP55 mRNA and protein was significantly decreased in the hsa-miR-195-5p agomir group. CEP55 level in hsa-miR-195-5p agomir+oe-E2F7 group and hsa-miR-195-5p agomir+oe-CEP55 group was prominently elevated and returned to the level of NC-agomir+oe-NC group (Fig. 7D–E). We, therefore, concluded that hsa-miR-195-5p/E2F7/CEP55 axis played a role in inhibiting LUAD tumor growth in vivo.
Hsa-miR-195-5p/E2F7/CEP55 axis can inhibit the growth of LUAD tumors. A–C Representative images (A), of tumors derived from xenograft animal models (B), tumor volumes (C), tumor weights; D, E The expression of CEP55 mRNA and protein in different mouse models. **, ***, **** meant P < 0.01, 0.001, 0.0001, respectively (one-way ANOVA)
Discussion
In this work, we investigated the role and regulatory mechanism of E2F7 in LUAD progression. Our results demonstrated a substantially high E2F7 level in LUAD. Additionally, silencing E2F7 inhibited autophagy of LUAD cells and promoted cell apoptosis and gemcitabine chemotherapy sensitivity by LUAD cells. Bioinformatics analysis revealed that E2F7 binds to TSS region of CEP55 promoter, and hsa-miR-195-5p binds to E2F7 3'-UTR region. Further studies demonstrated that hsa-miR-195-5p promoted apoptosis of LUAD cells via E2F7/CEP55 and inhibited autophagy and gemcitabine chemotherapy resistance of LUAD.
E2F family is the genome encoding various transcription factors in higher eukaryotes, and they participated in cell cycle, angiogenesis, DNA damage response, etc. (DeGregori and Johnson 2006; Endo-Munoz et al. 2009; Yang et al. 2020a). E2F7 stimulates metastasis and cell proliferation of colon cancer (Guo et al. 2020), esophageal cancer (Lu et al. 2020), and thyroid cancer (Guo and Zhang 2019). Our results suggested a significant up-regulation of E2F7 in LUAD tissues and cells. Previous investigations reported that E2F7 drives cancer development through the proliferation and differentiation of cancer cells in cutaneous squamous cell carcinoma and gallbladder carcinoma by competitively binding with E2F1 (Endo-Munoz et al. 2009; Xiang et al. 2019). In addition, in the studies about LUAD, it was manifested that E2F7 is target of miRNA-26a-5p (Liang et al. 2018) and miRNA-140-3p (Wang et al. 2021) and could promote the malignant progression of tumor cells. In our study, we found that there was a close relationship between E2F7 expression and proliferation and apoptosis in LUAD cells and that E2F7 functioned as an oncogene in LUAD progression.
The bioinformatics analysis unveiled that CEP55 is a target downstream of transcription factor E2F7. CEP55 has originally described as a key centrosome-related protein in mitotic outlet and cytokinesis (Fabbro et al. 2005). It has been reported that the up-regulation of CEP55 expression can facilitate cell migration and invasion and that high expression of CEP55 is implicated in poor prognoses of patients with liver cancer (Yang et al. 2020b), anaplastic thyroid cancer (Li et al. 2020), cervical cancer (Qi et al. 2018), and LUAD (Wu et al. 2019). Liu et al. (Liu et al. 2016) found that inhibition of CEP55 expression reduced viability and induced apoptosis of LUAD cells, which was consistent with the results of this study. We also verified the binding between CEP55 and E2F7 by using ChIP and dual-luciferase assays, and confirmed that E2F7 increased the activity of LUAD cells and inhibited apoptosis via activating CEP55 by using cell experiments.
Recently, a lot of studies confirmed the importance of autophagy in genesis, development, and treatment of tumors. Especially in the process of tumor treatment, a variety of treatments including radiotherapy and chemotherapy can lead to autophagy of tumor cells (Chiu et al. 2016; Michaud, et al. 2011). Autophagy is a protective mechanism that mediates drug resistance of tumor cells. Therefore, inhibiting autophagy re-sensitizes tumor cells and strengthen cytotoxicity of chemotherapy agents (Sui, et al. 2013). Autophagy is pivotal in the modulation of chemotherapy sensitivity of colon cancer, osteosarcoma, glioblastoma, and other malignant tumors (Lv et al. 2016; Xu et al. 2016; Li et al. 2018). Yin et al. (Lv et al. 2016; Xu et al. 2016; Li et al. 2018) found in a study of bladder cancer that CYLD overexpression and Livin gene knockout can improve gemcitabine chemotherapy sensitivity by reducing autophagy and increasing apoptosis. MiRNA-29c elevates gemcitabine chemotherapy sensitivity through inhibition of USP22-mediated autophagy of pancreatic cancer cells (Huang et al. 2018). We displayed that E2F7 level was closely related to autophagy and gemcitabine chemotherapy sensitivity of LUAD cells. Decreased E2F7 expression led to decreased autophagy level of LUAD cells, which in turn increased the sensitivity of LUAD cells to gemcitabine chemotherapy.
MiRNAs are endogenously expressed non-coding RNAs that can be used as modulators of post-transcriptional genes (Duan, et al. 2022). After identifying the regulatory mechanism of E2F7/CEP55 in LUAD, we continued to explore the upstream regulatory genes of E2F7. MiR-195-5p exerts an inhibitory role in the progression of various cancers, such as cervical cancer (Liu et al. 2020), lung cancer (Bu et al. 2021), colorectal cancer (Li et al. 2021), etc. Cheng et al. (Yuan et al. 2021) showed that miR-195-5p represses growth and promotes apoptosis of LUAD cells through targeting HOXA10 and enhances sensitivity of LUAD cells to X-ray irradiation. Herein, we reported that hsa-miR-195-5p was an upstream regulatory gene of E2F7, which could stimulate LUAD cell apoptosis, constrain proliferation and autophagy of LUAD cells, and enhance sensitivity of LUAD cells to gemcitabine via targeting E2F7/CEP55, which was basically in line with findings by Cheng et al. In a word, aberrant expression of hsa-miR-195-5p/E2F7/CEP55 can affect tumor cell autophagy, which may be a key pathway affecting drug resistance. In addition, Xu et al. (Xu et al. 2015) showed in their study on hepatocellular carcinoma that hsa-miR-195-5p significantly inhibits xenograft tumor growth in nude mice. In this study, xenograft experiments illustrated that hsa-miR-195-5p constrained LUAD tumor growth by targeting E2F7 and regulating CEP55, which was in line with in vitro results, again proving the reliability of the conclusions of this study.
To sum up, this research confirmed that hsa-miR-195-5p/E2F7/CEP55 axis is involved in the molecular mechanism of regulating autophagy and gemcitabine resistance of LUAD cells. The results of this study preliminarily confirmed that E2F7 may be an autophagy-related drug-resistance gene, providing a reference for clinical cancer treatment guided by the next-generation sequencing. However, there are still some deficiencies in this study. There was no in-depth investigation at the clinical level, which was a deficiency. We will further explore at the clinical level, so as to lay a more reliable basis for the precise treatment of LUAD.
Data Availability
The datasets generated and analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
References
An Q, Zhou L, Xu N (2018) Long noncoding RNA FOXD2-AS1 accelerates the gemcitabine-resistance of bladder cancer by sponging miR-143. Biomed Pharmacother 103:415–420. https://doi.org/10.1016/j.biopha.2018.03.138
Booth LA, Roberts JL, Dent P (2020) The role of cell signaling in the crosstalk between autophagy and apoptosis in the regulation of tumor cell survival in response to sorafenib and neratinib. Semin Cancer Biol 66:129–139. https://doi.org/10.1016/j.semcancer.2019.10.013
Bu L, Tian Y, Wen H, Jia W, Yang S (2021) miR-195-5p exerts tumor-suppressive functions in human lung cancer cells through targeting TrxR2. Acta Biochim Biophys Sin 53:189–200. https://doi.org/10.1093/abbs/gmaa159
Chen W et al (2021) Downregulation of lncRNA ZFAS1 inhibits the hallmarks of thyroid carcinoma via the regulation of miR3023p on cyclin D1. Mol Med Rep. https://doi.org/10.3892/mmr.2020.11640
Chiu HW et al (2016) Combination of the novel histone deacetylase inhibitor YCW1 and radiation induces autophagic cell death through the downregulation of BNIP3 in triple-negative breast cancer cells in vitro and in an orthotopic mouse model. Mol Cancer 15:46. https://doi.org/10.1186/s12943-016-0531-5
DeGregori J, Johnson DG (2006) Distinct and overlapping roles for E2F family members in transcription. Prolif Apoptosis Curr Mol Med 6:739–748. https://doi.org/10.2174/1566524010606070739
Duan L et al (2022) Immune-related miRNA-195–5p inhibits the progression of lung adenocarcinoma by targeting polypyrimidine tract-binding protein 1. Front Oncol 12:862564. https://doi.org/10.3389/fonc.2022.862564
Endo-Munoz L et al (2009) E2F7 can regulate proliferation, differentiation, and apoptotic responses in human keratinocytes: implications for cutaneous squamous cell carcinoma formation. Cancer Res 69:1800–1808. https://doi.org/10.1158/0008-5472.CAN-08-2725
Fabbro M et al (2005) Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell 9:477–488. https://doi.org/10.1016/j.devcel.2005.09.003
Guo H, Zhang L (2019) MicroRNA-30a suppresses papillary thyroid cancer cell proliferation, migration and invasion by directly targeting E2F7. Exp Ther Med 18:209–215. https://doi.org/10.3892/etm.2019.7532
Guo X, Liu L, Zhang Q, Yang W, Zhang Y (2020) E2F7 transcriptionally inhibits MicroRNA-199b expression to promote USP47, thereby enhancing colon cancer tumor stem cell activity and promoting the occurrence of colon cancer. Front Oncol 10:565449. https://doi.org/10.3389/fonc.2020.565449
He J et al (2015) Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy 11:373–384. https://doi.org/10.1080/15548627.2015.1009781
Huang L et al (2018) MicroRNA-29c increases the chemosensitivity of pancreatic cancer cells by inhibiting USP22 mediated autophagy. Cell Physiol Biochem 47:747–758. https://doi.org/10.1159/000490027
Levy JMM, Towers CG, Thorburn A (2017) Targeting autophagy in cancer. Nat Rev Cancer 17:528–542. https://doi.org/10.1038/nrc.2017.53
Li YJ et al (2017) Autophagy and multidrug resistance in cancer. Chin J Cancer 36:52. https://doi.org/10.1186/s40880-017-0219-2
Li H et al (2018) miR-519a enhances chemosensitivity and promotes autophagy in glioblastoma by targeting STAT3/Bcl2 signaling pathway. J Hematol Oncol 11:70. https://doi.org/10.1186/s13045-018-0618-0
Li YH, Xu KC, Huang GM, Zang HL (2020) The function and molecular mechanism of CEP55 in anaplastic thyroid cancer. Eur Rev Med Pharmacol Sci 24:9549–9555. https://doi.org/10.26355/eurrev_202009_23040
Li Y, Zhu Z, Hou X, Sun Y (2021) LncRNA AFAP1-AS1 promotes the progression of colorectal cancer through miR-195-5p and WISP1. J Oncol 2021:6242798. https://doi.org/10.1155/2021/6242798
Liang R et al (2018) SNHG6 functions as a competing endogenous RNA to regulate E2F7 expression by sponging miR-26a-5p in lung adenocarcinoma. Biomed Pharmacother 107:1434–1446. https://doi.org/10.1016/j.biopha.2018.08.099
Liu L, Mei Q, Zhao J, Dai Y, Fu Q (2016) Suppression of CEP55 reduces cell viability and induces apoptosis in human lung cancer. Oncol Rep 36:1939–1945. https://doi.org/10.3892/or.2016.5059
Liu X et al (2020) MiR-195-5p inhibits malignant progression of cervical cancer by targeting YAP1. Onco Targets Ther 13:931–944. https://doi.org/10.2147/ott.S227826
Lu T, Wang R, Cai H, Cui Y (2020) Long non-coding RNA DLEU2 promotes the progression of esophageal cancer through miR-30e-5p/E2F7 axis. Biomed Pharmacother 123:109650. https://doi.org/10.1016/j.biopha.2019.109650
Lv L et al (2016) Upregulation of CD44v6 contributes to acquired chemoresistance via the modulation of autophagy in colon cancer SW480 cells. Tumour Biol 37:8811–8824. https://doi.org/10.1007/s13277-015-4755-6
Ma T et al (2018) USP9X inhibition improves gemcitabine sensitivity in pancreatic cancer by inhibiting autophagy. Cancer Lett 436:129–138. https://doi.org/10.1016/j.canlet.2018.08.010
Michaud M et al (2011) Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science (New York) 334:1573–1577. https://doi.org/10.1126/science.1208347
Mulcahy Levy JM, Thorburn A (2020) Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ 27:843–857. https://doi.org/10.1038/s41418-019-0474-7
Qi J, Liu G, Wang F (2018) High levels of centrosomal protein 55 expression is associated with poor clinical prognosis in patients with cervical cancer. Oncol Lett 15:9347–9352. https://doi.org/10.3892/ol.2018.8448
Saha S, Panigrahi DP, Patil S, Bhutia SK (2018) Autophagy in health and disease: A comprehensive review. Biomed Pharmacother 104:485–495. https://doi.org/10.1016/j.biopha.2018.05.007
Sui X et al (2013) Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis 4:e838. https://doi.org/10.1038/cddis.2013.350
Tian S, Guo X, Yu C, Sun C, Jiang J (2017) miR-138-5p suppresses autophagy in pancreatic cancer by targeting SIRT1. Oncotarget 8:11071–11082. https://doi.org/10.18632/oncotarget.14360
Wang M et al (2018) Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis 9:1149. https://doi.org/10.1038/s41419-018-1187-4
Wang Y et al (2021) Circ-AASDH functions as the progression of early stage lung adenocarcinoma by targeting miR-140-3p to activate E2F7 expression. Transl Lung Cancer Res 10:57–70. https://doi.org/10.21037/tlcr-20-1062
Wu T et al (2015) Autophagy facilitates lung adenocarcinoma resistance to cisplatin treatment by activation of AMPK/mTOR signaling pathway. Drug Des Dev Ther 9:6421–6431. https://doi.org/10.2147/dddt.S95606
Wu S et al (2019) Correlation between EZH2 and CEP55 and lung adenocarcinoma prognosis. Pathol Res Pract 215:292–301. https://doi.org/10.1016/j.prp.2018.11.016
Xiang S et al (2019) E2F1 and E2F7 differentially regulate KPNA2 to promote the development of gallbladder cancer. Oncogene 38:1269–1281. https://doi.org/10.1038/s41388-018-0494-7
Xu H et al (2015) MicroRNA-195-5p acts as an anti-oncogene by targeting PHF19 in hepatocellular carcinoma. Oncol Rep 34:175–182. https://doi.org/10.3892/or.2015.3957
Xu R, Liu S, Chen H, Lao L (2016) MicroRNA-30a downregulation contributes to chemoresistance of osteosarcoma cells through activating Beclin-1-mediated autophagy. Oncol Rep 35:1757–1763. https://doi.org/10.3892/or.2015.4497
Yang Y, Klionsky DJ (2020) Autophagy and disease: unanswered questions. Cell Death Differ 27:858–871. https://doi.org/10.1038/s41418-019-0480-9
Yang R et al (2020a) E2F7-EZH2 axis regulates PTEN/AKT/mTOR signalling and glioblastoma progression. Br J Cancer 123:1445–1455. https://doi.org/10.1038/s41416-020-01032-y
Yang L, He Y, Zhang Z, Wang W (2020b) Upregulation of CEP55 predicts dismal prognosis in patients with liver cancer. Biomed Res Int 2020:4139320. https://doi.org/10.1155/2020/4139320
Ye X et al (2020) TNNC1 reduced gemcitabine sensitivity of nonsmall-cell lung cancer by increasing autophagy. Med Sci Monit 26:e922703. https://doi.org/10.12659/msm.922703
Yuan C et al (2021) Effects of MicroRNA-195-5p on biological behaviors and radiosensitivity of lung adenocarcinoma cells via targeting HOXA10. Oxid Med Cell Longev 2021:4522210. https://doi.org/10.1155/2021/4522210
Zhang M et al (2019) SOCS5 inhibition induces autophagy to impair metastasis in hepatocellular carcinoma cells via the PI3K/Akt/mTOR pathway. Cell Death Dis 10:612. https://doi.org/10.1038/s41419-019-1856-y
Zhao H, Ding F, Zheng G (2020) LncRNA TMPO-AS1 promotes LCN2 transcriptional activity and exerts oncogenic functions in ovarian cancer. FASEB J 34:11382–11394. https://doi.org/10.1096/fj.201902683R
Zhao Y, Zhang W, Yang Y, Dai E, Bai Y (2021) Diagnostic and prognostic value of microRNA-2355-3p and contribution to the progression in lung adenocarcinoma. Bioengineered 12:4747–4756. https://doi.org/10.1080/21655979.2021.1952367
Funding
This study was supported in part by grants from to investigate the value and mechanism of CEP55 gene as a diagnostic and independent prognostic factor in LUAD (2021RC033).
Author information
Authors and Affiliations
Contributions
Conception and design: LF. Administrative support: ZL. Provision of study materials or patients: LD. Collection and assembly of data: TZ. Data analysis and interpretation: JD. Manuscript writing: All authors. Final approval of manuscript: All authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors report no conflict of interest.
Ethical Approval
The study was approved by Shaoxing People’s Hospital Experimental Animal Ethics Committee, approval number [2022Z052]. The methods were carried out in accordance with the approved guidelines.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

10528_2023_10330_MOESM1_ESM.jpg
Supplementary file1 (JPG 1758 KB)—Supplementary Fig. 1 Effect of E2F7 knockdown/CEP55 overexpression on the functions of LUAD cells. A, B The mRNA and protein expression of E2F7 of HCC827 cells in each group; C The proliferation ability of each transfected group; D The apoptosis of HCC827 cells in each group; E The expression of apoptosis-related proteins in LUAD cells; F Immunofluorescence assay of E2F7 and LC3 protein expression in HCC827 cells in each group; G The expression of autophagy-related proteins in HCC827 cells; H, I The cell viability and IC50 values respond to 24 h and 48 h of gemcitabine treatment (0, 0.001, 0.01, 0.1, 1, 5, 10 μg/mL), respectively. *, **, ***, **** meant P<0.05, 0.01, 0.001, 0.0001, respectively (one-way ANOVA)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fu, L., Li, Z., Wu, Y. et al. Hsa-miR-195-5p Inhibits Autophagy and Gemcitabine Resistance of Lung Adenocarcinoma Cells via E2F7/CEP55. Biochem Genet 61, 1528–1547 (2023). https://doi.org/10.1007/s10528-023-10330-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10330-y