Abstract
Although many studies have investigated the association of paraoxonase 1 (PON1) polymorphisms with coronary artery disease (CAD). However, the outcomes were not consistent and remain uncertain. Therefore, it is the need of the hour to analyze the available literature and evaluate the association of PON1 polymorphisms with the CAD. All the relevant studies published in the English language from January 1, 2000, up to September 20, 2020, were identified by searching through various electronic databases. The two researchers independently extracted the information. The data were analyzed by using the MetaGenyo program. The pooled odds ratio was used to find the associations between CAD and PON1 polymorphisms. In the final analysis, we include 10 studies regarding the association of PON1 polymorphisms (rs662 and rs854560) with CAD. Overall, the Q192R polymorphism increased the risk of CAD in the tested genetic models including the homozygote model: OR 1.35, CI 1.02–1.79; allelic model: OR 1.16, CI 1.00–1.33; dominant model: OR 1.25, CI 1.03–1.52. The L55M polymorphism does not significantly associated with CAD in all the tested genetic models including the homozygote model: OR 1.00 CI, 0.64–1.56; allelic model: OR 1.02, 95% CI 0.84–1.23; dominant model: OR 1.08, CI 0.89–1.31. Further analysis showed no publication bias exists in meta-analysis. Our findings suggested that rs662 in the coding region was significantly associated with the CAD however, rs854560 has no significant association with the disease. Nevertheless, in future, there is a need for more studies with a larger sample size which may provide a more definite conclusion.
Study Registration: PROSPERO registration number CRD42020202278
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cardiovascular diseases (CVDs) are the group of disorders in the two systems, the circulatory system and the heart. Among the CVDs, the coronary artery disease (CAD) is the most prevalent. Due to the atherosclerosis the constriction of blood vessels leads to the poor blood supply to the heart that’s why coronary artery disease is also known as ischemic heart disease (IHD) (Ashiq et al. 2020). Clinical manifestations of coronary artery disease consist a host of chronic and acute conditions such as heart failure, stable angina and acute coronary syndrome (Shabana et al. 2018). In developing countries, its prevalence rate ranked at the top. In the South Asian countries the increasing prevalence of coronary artery disease poses a major threat and burden to the healthcare system (Jamee Shahwan et al. 2019). Worldwide, CVDs are responsible for 31% of all global mortality. Approximately 7.4 million deaths were reported due to the coronary artery disease (Shahid et al. 2018). According to the recent World Health Organization (WHO) report among the CVDs group the CAD represent a non-communicable killer in Pakistan (Larifla et al. 2016).
The coronary artery disease is a multifactorial disorder involving the complex interaction between the genetic and environmental factors (Sekhri et al. 2014). There are two broad categories of the conventional risk factors modifiable and non-modifiable risk factors. Obesity, hypertension, diabetes mellitus and smoking are the modifiable risk factors while family history, age and sex are the non-modifiable risk factors (Matam et al. 2015).
The human paraoxonase 1 (PON1) is a calcium dependent glycoprotein which have a molecular weight of 44 kDa. Its initial identification was done by its ability to hydrolyze multiple organophosphates, including sarin, soman, paraoxon and diazoxon and arylesters (Hassan et al. 2013). It predominantly expressed in the liver, which is a high-density lipoprotein associated esterase that hydrolyses lipoperoxides. It prevents the low density lipoprotein cholesterol concentration by hydrolyzing lipid peroxides. The enzymatic activity of paraoxonase1 enzyme varies among the individuals. The PON1 levels in serum are low if the high density lipoprotein levels are also low. There are several evidences of low PON1 activity in serum in the patients with lipid disorders such as diabetes mellitus, myocardial infarction, atherosclerosis and familial hypercholesterolemia (Taşkıran et al. 2009).
To date, up to 184 single nucleotide polymorphisms (SNPs) have been reported five common SNPs, two within the coding sequence Q192R [rs662], L55M [rs854560] region, five within the promoter and one seventy six within gene sequence (Gupta et al. 2012).
We selected two single nucleotide polymorphisms in the PON1 gene. The rs854560 caused by the substitution of the adenine at the place of thymine thus resulting M/L polymorphism at the position 55 (Shahsavari et al. 2020). In the rs662 caused by the substitution of the guanine at the place of the adenine which is located in exon six of the PON1 gene thus resulting Q/R SNP at 192 in protein. The SNPs in the PON1 gene affects the catalytic activity for various substrate hydrolysis and the oxidation of low density lipoprotein thus causing an increase in susceptibility to coronary artery disease (Ashiq et al. 2020).
Rationale
The results of rs662 and rs854560 in PON1 gene inconsistent among different ethnic groups, including Asian (Ahmad et al. 2012), Turkish (Hazar et al. 2011) and Caucasian population (Szpakowicz et al. 2016) thus it is the need of hour to analyze the available literature that provides us the most conclusive results for the association of these two SNPs with CAD.
Objectives
The aim of the current study was to evaluate the association of genetic polymorphisms in PON1 gene with coronary artery disease.
Materials and Methods
In the present study, we followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses 2009 statement. The current study is registered with PROSPERO (PROSPERO registration number CRD42020202278). A PRISMA checklist is shown in Table 1.
Search Strategy
We searched all published articles in, PubMed, MEDLINE, EMBASE, Web of Science, Ovid, and the Cochrane Library (from January 1, 2000, up to September 20, 2020) using the following MeSH terms and keywords:, including ‘coronary artery disease’, ‘CAD’, ‘PON1’, ‘Paraoxonase 1’, ‘gene polymorphism’, ‘variant’, ‘genotype’, ‘mutation’, ‘atherosclerosis’, ‘coronary heart disease’ and ‘worldwide’. To avoid missing any relevant study, we have also searched the related articles manually. Finally, all the duplicates studies were not included in the final analysis.
Inclusion and Exclusion Criteria
The articles consider eligible for the inclusion when the following conditions were met (1) retrospective case–control studies using either a hospital based design or population based design (2) The full length original articles on the association of PON1 polymorphisms and CAD in human subjects (3) Adequate information was provided for estimating the statistical analysis, including odds ratio (OR). Those not written in English language or not provided the adequate data and not designed as case–control studies or designed as meta-analysis or systematic reviews, were excluded.
Data Extraction
To reduce the selection bias, the authors used, predesigned data extraction table. Both the authors independently reviewed and extracted the data required from all the eligible studies. From each original article, the following data were abstracted: author names, year of publication, country, baseline characteristics, sample size (controls and cases), method, the distribution of genotypes and alleles in subjects, and evidence of conforming to the Hardy–Weinberg equilibrium (HWE). The detailed characteristics of each included study are given in Tables 2 and 3.
Quality Score Assessment
The quality of each included study was assessed by using the Newcastle–Ottawa Scale (NOS). The NOS ranges between 0 (worst) and 9 stars (best). Both the authors independently assessed the quality of articles and any disagreements were decided through discussion to achieve a consensus.
Statistical Analysis
The association of the PON1 polymorphism and risk of CAD was estimated by calculating the pooled ORs and 95%CI. The heterogeneity among the included studies was calculated using the chi-squared test and I2 statistic. A fixed effect model (Mantel–Haenszel) was used in the absence of heterogeneity and if heterogeneity is present then the random effect model (DerSimonian–Laird) was adopted to investigate the variation both from in-study and between-study. The genetic models used for both rs662 and rs854560 were: allelic model, homozygote model, recessive model and dominant model. The sensitivity test was performed to assess the stability of results. Publication bias was analyzed by using the funnel plot which was calculated by using the Begg’s and Egger’s tests. All statistical analysis was performed by using the MetaGenyo tool.
Results
Literature Search and Study Characteristics
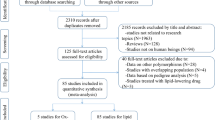
Through a literature searching, initially 365 published articles were identified, of which 355 articles were excluded as they did not investigate the association of PON1 polymorphisms with CAD. Thus, after a comprehensive literature search applying our inclusion criteria, 10 eligible full text published articles were included in the final analysis. The detailed screening process for all relevant literature is explained in a flow diagram shown in Fig. 1. Two studies involved Egyptian, Indian, 1 study involved Northern and Southern Han population and Iran whereas the other 4 studies were performed in European populations. Of all the 10 studies, controls of 5 studies were based on hospital population and the other five were from the general population (Table 2) (Godbole et al. 2020; Murillo-González et al. 2020; Fridman et al. 2016; Munshi et al. 2018; El-Lebedy et al. 2014; Birjmohun et al. 2009; Kaman et al. 2009; Liu et al. 2014; Mohamed et al. 2010; Shahsavari et al. 2020). The controls of 10 studies met Hardy–Weinberg equilibrium of genotype distributions. The detailed genotype distribution and allele frequencies were described in Table 3. All the included studies investigate the effect of rs662 and rs854560 genotypes on paraoxonase enzymatic activity (Table 4).
Association of rs662 with Coronary Artery Disease
The result of the pooled analysis of rs662 polymorphism in PON1 shows the significant association with coronary artery disease. We chose random effects models to merge all data based. Overall, the Q192R polymorphism increased the risk of CAD in all the tested genetic models (allelic model: OR 1.16, 95% CI 1.00–1.33; homozygote model: OR 1.35, CI 1.02–1.79; recessive model: OR 1.18, CI 0.94–1.49; dominant model: OR 1.25, CI 1.03–1.52). The main results of meta-analysis are shown in Fig. 2.
Association of rs854560 with Coronary Artery Disease
The result of the pooled analysis of rs854560 polymorphism in PON1 reveals the non- significant association with CAD. The main results of meta-analysis are shown in Fig. 3. We chose random-effects models to merge all data based. Overall, the L55M polymorphism does not increase the risk of CAD in all the tested genetic models (allelic model: OR 1.02, 95% CI 0.84–1.23; homozygote model: OR 1.00, CI 0.64–1.56; recessive model: OR 0.89 CI, 0.58–1.37; dominant model: OR 1.08, CI 0.89–1.31).
Sensitivity Analysis
Sensitivity analyses revealed that after excluding each literature the overall calculated odd ratio did not change significantly, which confirmed the stability and reliability of our analysis.
Publication Bias
The shapes of funnel plots do not show any apparent asymmetry in all the genetic models for both rs662 and rs854560 as shown in Figs. 4 and 5. The Eggers test also showed no statistical significant effect of the publication bias in both the tested SNPs. For the rs662 the p-value in all genetic models were non-significant for all genetic models (allelic: 0.77 homozygote: 0.57 recessive: 0.45 and dominant: 0.18). For the rs854560 the p-value in all genetic models were non-significant for all genetic models (allelic: 0.86 homozygote: 0.98 recessive: 0.88 and dominant: 0.75).
Discussion
Although several recent systematic review and meta-analysis, investigate the potential role of PON1 gene polymorphisms in coronary artery disease (Hernández-Díaz et al. 2016), but current analysis was the comprehensive assessment of its association with coronary artery disease. Moreover, our systematic review also provides insight about the effect of PON1 polymorphism on paraoxonase enzyme levels. The key characteristic of paraoxonase enzyme is that it exhibits hydrolytic activity and plays a vital role in inhibiting the oxidation of low density lipoprotein and high density lipoproteins. Thus, a single nucleotide polymorphism in the PON1 gene can affect the catalytic activity of enzyme and hence can be associated with CAD (Moreno-Godínez et al. 2018).
In the present study, we selected two major SNPs (rs662 and rs854560) in the PON1 gene and the pooled analysis results indicated a significant association between rs662 and coronary artery disease. While in the present study the rs854560 showed the non-significant association with CAD. The current study results for the SNP rs662 suggested that the minor allele G is significantly associated with coronary artery disease and the genotypes also have the significant effect on paraoxonase enzyme activity. We found the association between the CAD and all the genetic models tested for the rs662 in the analysis.
Our findings consistent with previous results reported by Shabana et al. (2018) and Liu et al. (2014) but contrast to those performed by the Birjmohun et al. (2009). Similarly a meta-analysis conducted by the Zhang et al. in Chinese population showed the weak association of Q192R polymorphism in PON1 with coronary artery disease (Zhang et al. 2018). Our study results also in general agreement those performed by the Kaur et al. in Asian Indians (Kaur et al. 2018). Lawlor et al. performed a meta-analysis in Caucasian populations, but they reported non-significant association of rs662 polymorphism in coronary artery disease (Lawlor et al. 2004). The results of the current study also in accordance to the meta-analysis study performed by the Wang et al. (Wang et al. 2018). Zeng et al. also performed a meta-analysis and reported the significant association of rs662 with CAD in Caucasians, South Asians and East Asians populations (Zeng and Zeng 2019).
We also found no association between our second SNP, i.e. rs854560 and coronary artery disease. The current analysis results consistent with the meta-analysis performed by the Hazar et al. in Turkish population (Hazar et al. 2011). While the significant association of L55M polymorphism reported by the Zeng and Zeng (2019). Similarly the meta-analysis results reported by the Hernández-Díaz et al. showed the significant association of the rs854560 polymorphism with CAD in European and Asian populations (Hernández-Díaz et al. 2016). Bounafaa et al. also reported that the M allele and R allele are significantly associated with coronary artery disease. Furthermore, these polymorphisms also affect the paraoxonase enzyme catalytic activity which may prove an important marker for monitoring the atherosclerosis (Bounafaa et al. 2015). In addition to all the analysis, it is also important to assess the publication bias that may introduce false positive in a meta-analysis study (Egger et al. 1997). So, in order to avoid this publication bias, we performed Begg and Egger test on the enrolled studies and we found no significant bias in our study. From the current analysis results, we confirmed that subjects with the R allele in PON1 gene are at high risk of CAD and need to be detected early in addition to receive the appropriate counseling in case contracting coronary artery disease.
Limitations
Though, the results of the present meta-analysis are quite comprehensive, however, there are a few study limitations also exist. First, we only select the articles that were published in English language and possibly this can influence the publication biasness. Second, in this study, we mainly focused on the two coding region SNPs in the PON1 gene and were not able to evaluate the other genes association with coronary artery disease.
Conclusions
It is concluded that the Q192R single nucleotide polymorphism in the coding region was significantly associated with CAD. However, in the current study L55M showed non-significant role in CAD. It is suggested that there is a need for more studies with a larger sample size in different subgroups, which could be beneficial to get a more definite conclusion.
References
Ahmad I, Narang R, Venkatraman A, Das N (2012) Two-and three-locus haplotypes of the paraoxonase (PON1) gene are associated with coronary artery disease in Asian Indians. Gene 506:242–247. https://doi.org/10.1016/j.gene.2012.06.031
Ashiq S, Ashiq K, Shabana S, Shahid SU, Qayyum M, Sadia H (2020) Prevalence and role of different risk factors with emphasis on genetics in development of pathophysiology of coronary artery disease (CAD). Pak Heart J 52:279–287
Birjmohun RS, Vergeer M, Stroes ES, Sandhu MS, Ricketts SL, Tanck MW, Wareham NJ, Jukema JW, Kastelein JJ, Khaw KT, Boekholdt SM (2009) Both paraoxonase-1 genotype and activity do not predict the risk of future coronary artery disease; the EPIC-Norfolk prospective population study. PLoS ONE 4:e6809. https://doi.org/10.1371/journal.pone.0006809
Bounafaa A, Berrougui H, Ghalim N, Nasser B, Bagri A, Moujahid A, Ikhlef S, Camponova P, Yamoul N, Simo OK, Essamadi A (2015) Association between Paraoxonase 1 (PON1) polymorphisms and the risk of acute coronary syndrome in a north African population. PLoS ONE 10:e0133719. https://doi.org/10.1371/journal.pone.0133719
Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634. https://doi.org/10.1136/bmj.315.7109.629
El-Lebedy D, Kafoury M, Abd-El Haleem D, Ibrahim A, Awadallah E, Ashmawy I (2014) Paraoxonase-1 gene Q192R and L55M polymorphisms and risk of cardiovascular disease in Egyptian patients with type 2 diabetes mellitus. J Diabetes Metab Disord 13:124. https://doi.org/10.1186/s40200-014-0125-y
Fridman O, Gariglio L, Riviere S, Porcile R, Fuchs A, Potenzoni M (2016) Paraoxonase 1 gene polymorphisms and enzyme activities in coronary artery disease and its relationship to serum lipids and glycemia. Arch Cardiol Mex 86:350–357. https://doi.org/10.1016/j.acmx.2016.08.001
Godbole C, Thaker S, Kerkar P, Nadkar M, Gogtay N, Thatte U (2020) Association of PON1 gene polymorphisms and enzymatic activity with risk of coronary artery disease. Future Cardiol. https://doi.org/10.2217/fca-2020-0028
Gupta N, Binu KB, Singh S, Maturu NV, Sharma YP, Bhansali A, Gill KD (2012) Low serum PON1 activity: an independent risk factor for coronary artery disease in North-West Indian type 2 diabetics. Gene 498:13–19. https://doi.org/10.1016/j.gene.2012.01.091
Hassan MA, Al-Attas OS, Hussain T, Al-Daghri NM, Alokail MS, Mohammed AK, Vinodson B (2013) The Q192R polymorphism of the paraoxonase 1 gene is a risk factor for coronary artery disease in Saudi subjects. Mol cell Biochem 380:121–128. https://doi.org/10.1007/s11010-013-1665-z
Hazar A, Dilmec F, Göz M, Koçarslan A, Aydin MS, Demirkol AH (2011) The paraoxonase 1 (PON1) gene polymorphisms in coronary artery disease in the southeastern Turkish population. Turk J Med Sci 41:895–902. https://doi.org/10.3906/sag-1009-1168
Hernández-Díaz Y, Tovilla-Zárate CA, Juárez-Rojop IE, González-Castro TB, Rodríguez-Pérez C, López-Narváez ML, Rodríguez-Pérez JM, Cámara-Álvarez JF (2016) Effects of paraoxonase 1 gene polymorphisms on heart diseases: systematic review and meta-analysis of 64 case-control studies. Medicine 95:e5298. https://doi.org/10.1097/MD.0000000000005298
Jamee Shahwan A, Abed Y, Desormais I, Magne J, Preux PM, Aboyans V, Lacroix P (2019) Epidemiology of coronary artery disease and stroke and associated risk factors in Gaza community–Palestine. PLoS ONE 14:e0211131. https://doi.org/10.1371/journal.pone.0211131
Kaman D, İlhan N, Metin K, Akbulut M, Üstündağ B (2009) A preliminary study of human paraoxonase and PON 1 L/M55–PON 1 Q/R 192 polymorphisms in Turkish patients with coronary artery disease. Cell Biochem Funct 27:88–92. https://doi.org/10.1002/cbf.1539
Kaur S, Bhatti GK, Vijayvergiya R, Singh P, Mastana SS, Tewari R, Bhatti JS (2018) Paraoxonase 1 gene polymorphisms (Q192R and L55M) are associated with coronary artery disease susceptibility in Asian Indians. Int J Diabetes Metab 21:38–47. https://doi.org/10.1159/000494508
Larifla L, Beaney KE, Foucan L, Bangou J, Michel CT, Martino J, Velayoudom-Cephise FL, Cooper JA, Humphries SE (2016) Influence of genetic risk factors on coronary heart disease occurrence in Afro-Caribbeans. Can J Cardiol 32:978–985. https://doi.org/10.1016/j.cjca.2016.01.004
Lawlor DA, Day IN, Gaunt TR, Hinks LJ, Briggs PJ, Kiessling M, Timpson N, Smith GD, Ebrahim S (2004) The association of the PON1 Q192R polymorphism with coronary heart disease: findings from the British Women’s Heart and Health cohort study and a meta-analysis. BMC Genet 5:17. https://doi.org/10.1186/1471-2156-5-17
Liu T, Zhang X, Zhang J, Liang Z, Cai W, Huang M, Yan C, Zhu Z, Han Y (2014) Association between PON1 rs662 polymorphism and coronary artery disease. Eur J Clin Nutr 68:1029–1035. https://doi.org/10.1038/ejcn.2014.105
Matam K, Khan IA, Hasan Q, Rao P (2015) Coronary artery disease and the frequencies of MTHFR and PON1 gene polymorphism studies in a varied population of Hyderabad, Telangana region in south India. J King Saud Univ Sci 27:143–150. https://doi.org/10.1016/j.jksus.2014.09.002
Mohamed RH, Mohamed RH, Karam RA, Abd El-Aziz TA (2010) The relationship between paraoxonase1-192 polymorphism and activity with coronary artery disease. Clin Biochem 43:553–558. https://doi.org/10.1016/j.clinbiochem.2009.12.015
Moreno-Godínez ME, Galarce-Sosa C, Cahua-Pablo JÁ, Rojas-García AE, Huerta-Beristain G, del Carmen A-R, Cruz M, Valladares-Salgado A, Antonio-Véjar V, Ramírez-Vargas MA, Flores-Alfaro E (2018) Genotypes of common polymorphisms in the PON1 gene associated with paraoxonase activity as cardiovascular risk factor. Arch Med Res 49:486–496. https://doi.org/10.1016/j.arcmed.2019.02.002
Munshi R, Panchal F, Chaurasia A, Rajadhyaksha G (2018) Association between paraoxonase 1 (PON1) Gene Polymorphisms and PON1 enzyme activity in Indian patients with coronary artery disease (CAD). Curr Pharmacogenom Pers Med 16:219–229. https://doi.org/10.2174/1875692117666181227112119
Murillo-González FE, Ponce-Ruiz N, Rojas-García AE, Rothenberg SJ, Bernal-Hernández YY, Cerda-Flores RM, Mackness M, Barrón-Vivanco BS, González-Arias CA, Ponce-Gallegos J, Medina-Díaz IM (2020) PON1 lactonase activity and its association with cardiovascular disease. Clin Chim Acta 500:47–53. https://doi.org/10.1016/j.cca.2019.09.016
Sekhri T, Kanwar RS, Wilfred R, Chugh P, Chhillar M, Aggarwal R, Sharma YK, Sethi J, Sundriyal J, Bhadra K, Singh S (2014) Prevalence of risk factors for coronary artery disease in an urban Indian population. BMJ Open 4:e005346. https://doi.org/10.1136/bmjopen-2014-005346
Shabana N, Ashiq S, Ijaz A, Khalid F, ul Saadat I, Khan K, Sarwar S, Shahid SU (2018) Genetic risk score (GRS) constructed from polymorphisms in the PON1, IL-6, ITGB3, and ALDH2 genes is associated with the risk of coronary artery disease in Pakistani subjects. Lipids Health Dis 17:224. https://doi.org/10.1186/s12944-018-0874-6
Shahid SU, Shabana NA, Rehman A, Humphries S (2018) GWAS implicated risk variants in different genes contribute additively to increase the risk of coronary artery disease (CAD) in the Pakistani subjects. Lipids Health Dis 17:89. https://doi.org/10.1186/s12944-018-0736-2
Shahsavari G, Nouryazdan N, Adibhesami G, Birjandi M (2020) Genetic associations and serum paraoxonase levels with atherosclerosis in western Iranian patients. Mol Biol Rep 47:5137–5144. https://doi.org/10.1007/s11033-020-05585-2
Szpakowicz A, Pepinski W, Waszkiewicz E, Maciorkowska D, Skawronska M, Niemcunowicz-Janica A, Dobrzycki S, Musial WJ, Kaminski KA (2016) The influence of renal function on the association of rs854560 polymorphism of paraoxonase 1 gene with long-term prognosis in patients after myocardial infarction. Heart Vessels 31:15–22. https://doi.org/10.1007/s00380-014-0574-8
Taşkıran P, Çam SF, Şekuri C, Tüzün N, Alioğlu E, Altıntaş N, Berdeli A (2009) The relationship between paraoxanase gene Leu-Met (55) and Gln-Arg (192) polymorphisms and coronary artery disease. Turk Kardiyol Dern Ars 37:473–478
Wang Y, Chen Y, Zhai X, Zhao X, Guo R, Yu B, Zhang W, Xie J (2018) Genetic variants of the paraoxonase 1 gene and risk of coronary heart disease and stroke in the Chinese population: a meta-analysis. Int J Clin Exp Med 11:3010–3022
Zeng Q, Zeng J (2019) A meta-analysis on relationship between paraoxonase 1 polymorphisms and atherosclerotic cardiovascular diseases. Life Sci 232:116646. https://doi.org/10.1016/j.lfs.2019.116646
Zhang Z, Ou J, Cai P, Niu B, Li J (2018) Association between the PON1 Q192R polymorphism and coronary heart disease in Chinese: a meta-analysis. Medicine 97:e11151. https://doi.org/10.1097/MD.0000000000011151
Funding
Not Applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ashiq, S., Ashiq, K. The Role of Paraoxonase 1 (PON1) Gene Polymorphisms in Coronary Artery Disease: A Systematic Review and Meta-Analysis. Biochem Genet 59, 919–939 (2021). https://doi.org/10.1007/s10528-021-10043-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-021-10043-0