Abstract
Ammonia can easily accumulate in water during the culture period, and the accumulated concentrations can reach high levels, which is one of the main stress factors with severely negative effects on the growth and health of cultured shrimp. Dietary manipulation, such as feeding Ampithoe sp. meal, has been proven to be an effective method for reducing the mortality of Litopenaeus vannamei under sublethal ammonia stress and thus improving the ammonia tolerance of the shrimp, but the underlying mechanism needs to be further studied. In this study, L. vannamei with increased ammonia tolerance (PV_T) were obtained by feeding Ampithoe sp. meal, shrimp not fed Ampithoe sp. meal were used as a control (PV_C), and a comprehensive transcriptome analysis of the response of L. vannamei exposed to ammonia-N at the 50% lethal concentration for 96 h was performed. A total of 318,815,732 clean reads, including 170,156,604 and 148,659,128 clean reads from the shrimp in the PV_T and PV_C groups, were obtained. The expression of genes involved in carbohydrate and lipid metabolism pathways was upregulated in the PV_T group compared with the PV_C group, which indicated that the shrimp in the PV_T group had a higher energy supply level than those in the PV_C group, and this difference was the main cause of the observed improvement in ammonia tolerance. Additionally, the upregulated expression of genes involved in the AMP-activated protein kinase (AMPK) pathway in the PV_T group was responsible for the observed upregulation of the expression of genes involved in the carbohydrate and lipid metabolism pathways. However, among the GO terms that were significantly enriched in the DEGs identified from the comparison of the PV_T and PV_C groups, only one GO term was related to the immune system process. In conclusion, feeding Ampithoe sp. meal to L. vannamei exposed to sublethal ammonia can enhance the carbohydrate and lipid metabolism pathways to increase the energy supply and thus reduce the mortality of the shrimp.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The Pacific white shrimp Litopenaeus vannamei is native to the Pacific coastal waters of South America. Due to its fast growth, strong adaptability, high protein content, and delicious taste, L. vannamei is currently the most important shrimp species worldwide, and its total world culture output is 4.16 million tons (FAO. 2018). In recent years, the density of shrimp culture has increased to obtain a higher yield, and the shrimp culture environment has exhibited gradual deterioration, which often causes shrimp to experience various environmental stresses. As a result, the morbidity and mortality of shrimp have increased, which has seriously reduced the efficiency and restricted the development of the shrimp culture industry (Romano and Zeng 2013).
Ammonia is one of the main stress factors affecting shrimp culture and is mainly derived from the degradation of nitrogen-containing organic matter in the culture environment, such as residual feed and shrimp excrement (Chatvijitkul et al. 2017). Ammonia exists in two forms in the environment: nonionic ammonia (NH3) and ionic ammonia (NH4+). NH3 easily diffuses into aquatic animals due to its small molecular diameter, which leads to a reduction in the oxygen-carrying capacity of blood and damage to the mucous membrane and nervous system (Romano and Zeng 2013). Therefore, NH3 exerts a stronger toxic effect on aquatic animals than NH4+. However, although the toxicity of NH4+ is relatively weak, high concentrations of NH4+ hinder the ammonia excretion process of aquatic animals, which results in the accumulation of ammonia in the organisms (Armstrong et al. 1978). The toxic effect of ammonia directly affects the normal growth of aquatic animals. High concentrations of ammonia reduce the growth rate of shrimp, increase the molting frequency and mortality, and thus cause great economic loss to the shrimp culture industry (Chen and Kou 1992; Miranda-Filho et al. 2009). A study conducted by Liang et al. (2016) indicated that exposure to ammonia at 20 mg/L could induce oxidative stress, which further induces endoplasmic reticulum stress and apoptosis in the hepatopancreas of L. vannamei. Studies have suggested that ammonia exposure inhibits the immune system of L. vannamei, which results in susceptibility to pathogens, such as Vibrio alginolyticus and the causative agent of white spot syndrome virus (Liu and Chen 2004; Kathyayani et al. 2019; Liu et al. 2020). A study conducted by Lin and Chen (2001) indicated that the 96-h 50% lethal concentration values (LC50) of ammonia-N and NH3-N on L. vannamei juveniles were 24.39 mg/L, 35.4 mg/L, and 39.54 mg/L and 1.20, 1.57, and 1.60 mg/L at salinities of 15‰, 25‰, and 35‰, respectively. The “safety level” for ammonia-N at salinities of 15‰, 25‰, and 35‰ was estimated to equal 2.44, 3.55, and 3.95 mg/L, respectively.
Previous studies have shown that dietary manipulation can regulate metabolic processes in aquatic animals and enhance their ability to resist stress (Mai et al. 2004; Martins et al. 2012; Zhang et al. 2012; Dawood et al. 2017). Natural feed organisms are rich in a variety of nutrients and active substances, which can be used as nutritional fortifiers in shrimp culture to enhance resistance to stress (Sirirustananun et al. 2011; Xiao et al. 2017). Ampithoe sp. is a type of nutritious natural feed organism that is rich in crude protein (51.2%), crude lipids (5.8%), and essential amino acids (23.4%) (Peng et al. 2015). Dietary supplementation with Ampithoe sp. meal can reduce the mortality of L. vannamei under sublethal ammonia stress, which indicates that the ammonia tolerance of the shrimp is improved (Shan et al. 2018); however, the underlying mechanism, which may involve the regulation of a series of physiological pathways, remains unclear. Revealing the mechanisms can provide targets for establishing methods for mitigating the effects of ammonia stress on cultured shrimp.
The ammonia tolerance of L. vannamei enhanced by Ampithoe sp. meal was described in our previous study, which indicated that shrimp with high ammonia tolerance exposed to a sublethal ammonia-N stress concentration of 68.00 mg/L (1.61 mg/L nonionic ammonia) exhibit higher levels of carbohydrate and lipid metabolism under ammonia stress (Wang et al. 2020a). In recent years, transcriptomic analyses have been performed to better understand the physiological functions and molecular biological processes of shrimp under ammonia stress (Lu et al. 2016; Li et al. 2018). Therefore, in the present study, to reveal the molecular mechanism underlying the improvement in the ammonia tolerance of shrimp achieved by dietary manipulation, L. vannamei with high ammonia tolerance (PV_T) was obtained by feeding Ampithoe sp. meal, and shrimp that were not fed Ampithoe sp. meal served as a control (PV_C). Subsequently, a comprehensive transcriptomic analysis of L. vannamei under sublethal ammonia stress was performed using Illumina sequencing and bioinformatics. The present study could provide important information for understanding the molecular mechanism underlying the improvements in the environmental tolerance of shrimp obtained by dietary manipulation, and this information provides targets for establishing methods involving dietary manipulation that could mitigate the effects of ammonia stress on cultured shrimp.
Materials and methods
Experimental diets
Ampithoe sp. was obtained from Changyi County, Weifang City, Shandong Province, China. Ampithoe sp. meal was prepared as described by Shan et al. (2018). Fish meal, gelatinized cornstarch, fish oil, and lecithin were used as the main raw materials used to make the feed; the nutritionally fortified diet contained 33% Ampithoe sp. meal, and no Ampithoe sp. meal was added to the control diet. The formula, preparation, and storage of the two diets were described by Wang et al. (2020a).
Experimental design
A previous study indicated that a diet manipulation period of 21 days is sufficient for improving the ammonia tolerance of shrimp (Wang et al. 2020a); therefore, 21 days was selected as the diet manipulation period in this study. The experiment was performed in an indoor water recycling system consisting of 50-L water tanks. L. vannamei was purchased from Rushan City, Shandong Province, China. The shrimp were first acclimated for 7 days, and shrimp of similar sizes (body length 6.5 ± 0.2 cm, weight 2.8 ± 0.2 g) were then randomly divided into the PV_C and PV_T groups for 21-day dietary manipulation. The shrimp in the PV_C group were fed the control diet, and those in the PV_T group were fed the nutritionally fortified diet. Each group contained thirteen repetitions (one water tank per repetition, 20 shrimp per repetition), and the shrimp were fed the corresponding diet at 07:00, 10:00, 18:00, and 22:00 every day. The water quality indicators of the indoor water recycling system were as follows: water temperature of 26 ± 1 ℃, salinity of 30.0 ± 0.5‰, dissolved oxygen > 6 mg/L, and pH ranging from 7.6 to 7.8.
At the end of the 21-day dietary manipulation period, a 96-h ammonia stress test was performed. Six repetitions (20 shrimp per repetition) were included in each group; three of the repetitions were used to collect samples, and the three other repetitions were used to record the mortality. The body length and weight of the shrimp after 21 days of feeding are presented in Table S1, and no significant difference in the body length and weight of shrimp was found between the PV_C and PV_T groups. The 96-h LC50 of ammonia-N was 68.00 mg/L (1.61 mg/L nonionic ammonia), which was selected as the stress concentration, and NH4Cl was used to increase the concentration of ammonia-N used in this study according to a previous study (Wang et al. 2020a). The ammonia stress test was performed for 96 h in the present study. At the end of the ammonia stress test, nine shrimp were randomly selected from each group, and hepatopancreas tissue samples were collected and then stored at \(-\) 80 °C for transcriptome analyses.
In 2019, the shrimp genome was decoded (Zhang et al., 2019), which provided an important reference for related research on shrimp molecular biology. To improve the data from those obtained in previous studies, we reanalyzed the data for the comparative transcriptome analysis between the ammonia-challenged and control groups from the “ammonia-sensitive” family of L. vannamei published in the study by Lu et al. (2016) using the same approach as that used for the data in our study based on published genomics. A total of 239,706,580 raw reads were downloaded from the Short Read Archive of the National Center for Biotechnology Information (NCBI) with the accession number SRP062191. The data from the shrimp subjected to ammonia challenge were recorded as the S1 group, and those from the shrimp without ammonia challenge were recorded as the S0 group.
RNA isolation and Illumina sequencing
The nine hepatopancreas tissue samples that were collected at the end of the ammonia stress test from each group were divided into three replicates (each replicate contained three samples). Total RNA was extracted from each replicate using the RNAiso Plus Kit (TaKaRa, Japan) according to the manufacturer’s instructions. The concentration and purity of the RNA were detected using a NanoDrop 2000 spectrophotometer. The RNA integrity was detected by agarose gel electrophoresis, and the RIN value was determined with an Agilent 2100 instrument to ensure the high quality of the RNA (total RNA ≥ 2 μg, OD260/280 = 1.8 ~ 2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5, 28S:18S ≥ 1.0).
The RNA library was established using the TruSeq™ RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Total RNA was extracted and used for mRNA enrichment using magnetic beads with oligo (dT) to analyze the transcriptome information. The mRNA was randomly fragmented into approximately 200-bp fragments using fragmentation buffer and reverse transcribed into first-strand cDNA by the addition of random primers using the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen, CA, USA). Second-strand cDNA was synthesized by adding DNA polymerase I, RNase H, dNTPs, and buffer. The cDNA fragment was then purified using the QIAquick PCR Extraction Kit and end repaired, and poly (A) was subsequently added. These fragments were then ligated with sequencing adapters. Fragments of suitable length were obtained and subjected to 15 cycles of amplification. The PCR products were then purified, assessed, and sequenced on the Illumina HiSeq™ 2000 platform to generate the raw reads.
Bioinformatic analysis
The raw sequencing data generated in this study have been submitted to the NCBI with the accession number PRJNA694351. The raw reads were transformed into clean reads by removing the adapter sequences, ribosomal RNA (rRNA), low-quality reads, and empty reads. The treatments were performed through SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickl) (Ma et al. 2016). The clean reads were mapped to the rRNA database using Bowtie2 (Langmead and Salzberg 2012), and the rRNA was removed. The clean reads were then aligned to the L. vannamei genome (NCBI accession number: ASM378908v1) (Wang et al. 2020b) using HISAT2 (Kim et al. 2015), and expression values were estimated based on alignment using StringTie (Pertea et al. 2015). The edgeR package (http://www.r-project.org/) was used to identify the significantly differentially expressed genes (DEGs) between the two groups. The genes with a corrected p value < 0.05 and |log2 (fold change)|≥ 1 identified from the comparison were identified as DEGs. The DEGs were then subjected to Gene Ontology (GO) functional enrichment analysis using Goatools (Klopfenstein et al. 2018), and an enrichment analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways based on the DEGs was performed with an R language script and Fisher’s exact test.
qPCR analysis of selected genes from the transcriptome
To validate the expression profile data, eight genes were selected randomly to quantify their relative expression levels by quantitative real-time PCR (qPCR). The housekeeping gene encoding β-actin was used as an internal standard. The primer sequences of the genes are presented in Table 1. Total RNA was isolated using RNAiso Plus (TaKaRa, Japan) according to the manufacturer’s instructions and treated with the PrimeScript™ RT Reagent Kit (TaKaRa, Japan) for the synthesis of first-strand cDNA. PCR amplification was performed under the following conditions: 95 °C for 10 s and 40 cycles of 95 °C for 10 s and 60 °C for 30 s. At the end of the reaction, the reaction specificity was detected by melting curve analysis. The 2−ΔΔ CT method was used to analyze the relative expression levels of different genes.
Statistical analysis
The data obtained from the qPCR analysis and cumulative mortality assays were analyzed with SPSS version 25.0 for Windows. Independent samples t tests were performed to evaluate the final cumulative mortality and the relative expression levels of DEGs between the PV_C and PV_T groups. Differences with a p value < 0.05 were considered significant.
Results
Cumulative mortality
The cumulative mortality observed during the ammonia stress test is presented in Fig. 1. The shrimp in the PV_C group began to die after 6 h of ammonia stress, whereas those in the PV_T group began to die after 36 h. At the end of the ammonia stress test, although no significant difference was found between the groups (p = 0.189), the cumulative mortality of the PV_C and PV_T groups was 46.7% and 30.0%, respectively. The cumulative mortality of the PV_T group was decreased by 35.7% compared with that of the PV_C group.
Illumina sequencing and read mapping
A total of 322,357,856 raw reads were generated from the PV_C and PV_T groups. After removing the raw reads with adapter sequences, ambiguous nucleotides, and contamination, a total of 318,815,732 clean reads were generated, and these included 148,659,128 reads for the three PV_C libraries and 170,156,604 reads for the three PV_T libraries. More than 97% of the bases had a base accuracy of 99%, and more than 93% of the bases had a base accuracy of 99.9%. These clean reads were mapped against the annotated genome of L. vannamei, and more than 89% of the reads were successfully mapped (Table 2).
Differentially expressed genes
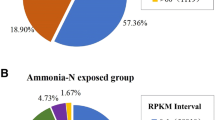
Comparative transcriptome analyses were performed between the PV_T and PV_C groups and between the S1 and S0 groups based on a threshold p value < 0.05. As shown in Fig. 2, a total of 1,778 DEGs were detected between PV_T and PV_C groups, and these DEGS included 749 significantly upregulated genes and 1,029 significantly downregulated genes. A total of 1,357 DEGs, including 991 significantly upregulated genes and 366 significantly downregulated genes, were detected between the S1 and S0 groups. Two hundred fifty DEGs were shared between the PV_C/PV_T and S0/S1 comparisons, and 16 of these genes showed the same changing trend. The p value, fold ratio, and annotation information of all the DEGs are provided in Supplementary Tables S2 and S3.
GO analysis
All annotated genes were divided into three categories: molecular function, cellular component, and biological process. The results showed that the DEGs obtained from the comparison of the PV_T and PV_C groups were significantly enriched in 182 GO terms (p value < 0.05) (Fig. 3 and Supplementary Table S4). Among these terms, the top three enriched terms in the biological process category were organic substance biosynthetic process (GO:1,901,576), metabolic process (GO:0,008,152), and biosynthetic process (GO:0,009,058).
KEGG pathway analysis
KEGG pathway analysis identifies molecular interaction networks within cells and helps elucidate the potential biological functions of analyzed genes. The major pathways associated with the DEGs between the PV_T and PV_C groups that were detected by KEGG enrichment analysis are presented in Fig. 4 and Table 3. As shown in Table 3, eight significantly enriched KEGG pathways were related to metabolism, and among these, oxidative phosphorylation, arachidonic acid metabolism, pentose and glucuronate interconversions, and pyruvate metabolism were the most frequently represented pathways in the response to ammonia stress.
Scatterplot showing the enrichment of KEGG pathways among the DEGs. The vertical axis represents the pathway categories, and the horizontal axis shows the enrichment factor. The point size is proportional to the number of DEGs in each pathway. The point colors correspond to different p values, as indicated on the right
Verification of transcriptome data by qPCR
In the present study, eight DEGs between the tolerance and control group were selected based on the functional and pathway enrichment results for qPCR validation of their expression patterns in the transcriptome data. The quantitative results were all well correlated with the transcriptome results (Fig. 5), which confirmed that the sequencing-based results were credible.
Discussion
Ammonia exerts negative effects on shrimp in the culture environment, and research on enhancing the ammonia tolerance of shrimp through dietary manipulation has been performed, but little information on the molecular mechanism underlying the improvements in ammonia tolerance achieved by dietary manipulation is available. This study reports the key pathways that respond to ammonia stress in shrimp with different degrees of ammonia tolerance caused by dietary manipulation, as determined by RNA-Seq technology, and the results suggest that the improvement in the ammonia tolerance of shrimp achieved by dietary manipulation may be mainly related to the metabolic process.
The present study indicated a delayed onset of death and markedly lower cumulative mortality in the PV_T group compared with the PV_C group under ammonia stress. This result confirmed that dietary manipulation could improve the ammonia tolerance of L. vannamei, which was consistent with the results of our previous studies (Shan et al. 2018, 2019).
Our previous studies have shown significant differences in energy metabolism levels among shrimp with different degrees of ammonia tolerance under ammonia stress. Shrimp with high ammonia tolerance exhibited higher activities of enzymes related to carbohydrate and lipid metabolism than those observed in the control shrimp, and these higher activities provide shrimp with more energy to cope with ammonia stress (Shan et al. 2019; Wang et al. 2020a). In the present study, a GO term enrichment analysis identified 426 DEGs between the PV_T and PV_C groups, and these DEGs were significantly enriched in 182 GO terms. In addition, 219 of these DEGs were significantly enriched in metabolic processes and were mainly related to carbohydrate metabolism and lipid metabolism: the carbohydrate metabolism pathways mainly included starch and sucrose metabolism, fructose and mannose metabolism, and the citrate cycle, and the lipid metabolism pathways included glycerolipid metabolism, linoleic acid metabolism, and arachidonic acid metabolism. Carbohydrates and lipids are the main energy supply substances in shrimp and provide energy to help shrimp cope with environmental stress (Sánchez-Paz et al. 2007; Chen et al. 2015). A study conducted by Racotta and Roberto (2000) found that exposure to a high concentration of ammonia could increase the energy expenditure of shrimp and diminish the ability of the hemolymph to transport oxygen, which may lead to a shortage of energy supply at the late stage of ammonia stress. In the present study, the expression of most DEGs in carbohydrate metabolism and lipid metabolism was significantly upregulated in the tolerance group compared with the control group, which indicated that feeding Ampithoe sp. meal could improve the energy supply capacity of L. vannamei under ammonia stress and thus improve the ammonia tolerance of the shrimp.
The analysis of DEGs between the S1 and S0 groups revealed that the expression of DEGs in the linoleic acid and arachidonic acid metabolism pathways was significantly downregulated by ammonia stress. A similar study conducted by Xiao et al. (2019) also found that the arachidonic acid metabolism pathway was significantly decreased, which indicated that lipid metabolism was disrupted in shrimp after ammonia exposure. Compared with carbohydrates, lipids were the most important energy supply substance in the response of shrimp to ammonia stress. Previous studies have confirmed that L. vannamei could reduce the catabolism of carbohydrates and increase that of lipids to meet the energy demand under ammonia stress (Racotta and Roberto 2000). A study conducted by Chen et al. (2014) also showed that L. vannamei needs to obtain additional energy from saturated fatty acids to ensure its normal physiological function under environmental stress. In this study, the disrupted lipid metabolism in shrimp belonging to the S1 group decreased the energy supply needed to cope with ammonia stress. However, the expression of DEGs involved in the linoleic acid and arachidonic acid metabolism pathways was significantly upregulated in the PV_T group compared with the PV_C group, and these findings indicate that feeding Ampithoe sp. meal could alleviate the damage to lipid metabolism caused by ammonia stress and plays a protective role in lipid metabolism, which further enhances the energy supply of L. vannamei in response to ammonia stress.
Studies have indicated that changes in environmental conditions would cause an increase in energy use to restore and maintain homeostasis in the body (Calow 1991; Calow and Forbes 1998). An insufficient energy supply under environmental stress severely affects the energy balance of the body. Energy imbalance can lead to disorder in bodily functions, which would damage the vitality and health of an organism (Tatar and Carey 1995; Honkoop 2003; Guerra et al. 2012). Therefore, energy homeostasis is of great significance for environmental tolerance. AMPK is a key regulator of energy balance in organisms that can regulate cellular energy metabolism by activating the ATP production pathway and blocking the ATP consumption pathway (Hardie 2003, 2008). The transcriptome analysis performed in this study revealed that the DEGs in the AMPK pathway showed significantly upregulated expression in the S1 group compared with the S0 group. The results of this study were consistent with those of other studies showing that the expression of AMPK in the hepatopancreas of L. vannamei is upregulated under acute stress to provide enough energy to cope with the stress (Xu et al. 2016; Han et al. 2013; Jost et al. 2012).
The transcriptome analysis of DEGs between the tolerance and control groups found that the transcription levels of three subunits of AMPK, namely, AMPK-α, AMPK-β, and AMPK-γ, were all increased in the hepatopancreas of shrimp in the tolerance group compared with the control group. All three subunits are related to energy metabolism (Hardie et al. 2003). Among these subunits, AMPK-α is a catalytic subunit (Hawley et al. 1996) that can increase energy consumption in the hepatopancreas of L. vannamei to ensure activity of digestive enzymes under stress (Xu et al. 2016). AMPK-β is a regulatory subunit that is closely related to the metabolism of carbohydrates (Hudson et al. 2003; Polekhina et al. 2003). AMPK-γ is also a regulatory subunit that can be combined with AMP and ATP and change the energy demand of the organism by switching on or off the ATP synthesis and catabolism pathways (Choi et al. 2001). The activated AMPK pathway could induce the catabolism pathway and decrease the pathway of lipid and carbohydrate synthesis to generate ATP and thus regulate the energy balance of the organism under stress (Hardie et al. 2012). The upregulation of AMPK expression in the hepatopancreas of shrimp in the PV_T group would increase the energy supply in the body, which might result in upregulation of the expression of DEGs in carbohydrate and lipid metabolism pathways, and this upregulation could increase the energy level of shrimp to cope with ammonia stress.
The GO term enrichment analysis of DEGs between the S1 and S0 groups indicated that the immune defense function of shrimp was affected by ammonia stress (Table S5). Lu et al. (2016) speculated that ammonia stress could reduce the immune defense ability of shrimp and increase susceptibility to pathogens. The study conducted by Xiao et al. (2019) also showed that ammonia stress could change the expression of immune-related genes in the hepatopancreas of L. vannamei and induce inhibition of the immune system. Other studies of the response of shrimp to ammonia stress have also focused on changes in immunity (Chen et al. 2012; Wongsasak et al. 2015; Duan et al. 2017). However, among the 36 GO terms that were significantly enriched in DEGs between PV_T and PV_C, only one GO term was related to the immune system process. This result suggested that feeding Ampithoe sp. meal did not affect immune processes in L. vannamei under ammonia stress.
Conclusion
Under ammonia stress, the expression of many genes and pathways in the hepatopancreas of L. vannamei with different levels of ammonia tolerance caused by dietary manipulation (feeding Ampithoe sp. meal) significantly differed, and these genes and pathways were mainly associated with metabolic processes. Genes involved in carbohydrate and lipid metabolism pathways showed upregulated expression in the PV_T group compared with the PV_C group, which indicated that an increased energy supply under ammonia stress resulted in increased energy mobilization to cope with stress. However, feeding Ampithoe sp. meal did not affect the immune process of shrimp under ammonia stress. This study provides important information for future studies on the mechanism of dietary manipulation and methods for improving the ammonia tolerance of cultured shrimp.
Data availability
Not applicable.
Code availability
Not applicable.
References
Armstrong DA, Chippendale D, Knight AW, Colt JE (1978) Interaction of ionized and un-ionized ammonia on short-term survival and growth of prawn larvae. Macrobrachium Rosenbergh Biol Bull-US 154(1):15–31. https://doi.org/10.2307/1540771
Calow P (1991) Physiological costs of combating chemical toxicants: ecological implications. Comp Biochem Phys C 100(1):3–6. https://doi.org/10.1016/0742-8413(91)90110-F
Calow P, Forbes VE (1998) How do physiological responses to stress translate into ecological and evolutionary processes? Comp Biochem Phys A 120(1):11–16. https://doi.org/10.1016/S1095-6433(98)10003-X
Chatvijitkul S, Boyd CE, Davis DA, McNevin AA (2017) Pollution potential indicators for feed-based fish and shrimp culture. Aquaculture 477:43–49. https://doi.org/10.1016/j.aquaculture.2017.04.034
Chen JC, Kou YZ (1992) Effects of ammonia on growth and molting of Penaeus japonicus juveniles. Aquaculture 104(3–4):249–260. https://doi.org/10.1016/0044-8486(92)90207-2
Chen YY, Sim SS, Chiew SL, Yeh ST, Liou CH, Chen JC (2012) Dietary administration of a Gracilaria tenuistipitata extract produces protective immunity of white shrimp Litopenaeus vannamei in response to ammonia stress. Aquaculture 370:26–31. https://doi.org/10.1016/j.aquaculture.2012.09.031
Chen K, Li EC, Gan L, Wang XD, Xu C, Lin HZ, Qin JG, Chen LQ (2014) Growth and lipid metabolism of the pacific white shrimp Litopenaeus vannamei at different salinities. J Shellfish Res 33(3):825–832. https://doi.org/10.2983/035.033.0317
Chen K, Li EC, Li TG, Xu C, Wang XD, Lin HZ, Qin JG, Chen LQ (2015) Transcriptome and molecular pathway analysis of the hepatopancreas in the Pacific white shrimp Litopenaeus vannamei under chronic low-salinity stress. PLoS ONE 10(7):e0131503. https://doi.org/10.1371/journal.pone.0131503
Choi SL, Kim SJ, Lee KT, Kim J, Mu J, Birnbaum MJ, Kim SS, Ha J (2001) The regulation of AMP-activated protein kinase by H2O2. Biochem Bioph Res Co 287(1):92–97. https://doi.org/10.1006/bbrc.2001.5544
Dawood MAO, Koshio S, Ishikawa M, Yokoyama S, El Basuini MF, Hossain MS, Nhu TH, Moss AS, Dossou S, Wei H (2017) Dietary supplementation of β-glucan improves growth performance, the innate immune response and stress resistance of red sea bream, Pagrus major. Aquacult Nutr 23:148–159. https://doi.org/10.1111/anu.12376
Duan YF, Zhang Y, Dong HB, Wang Y, Zheng XT, Zhang JS (2017) Effect of dietary Clostridium butyricum on growth, intestine health status and resistance to ammonia stress in Pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immun 65:25–33. https://doi.org/10.1016/j.fsi.2017.03.048
FAO, 2018. The state of world fisheries and aquaculture (SOFIA 2018) [Online]. FAO,
Guerra C, Zenteno-Savín T, Maeda-Martínez AN, Philipp EER, Abele D (2012) Changes in oxidative stress parameters in relation to age, growth and reproduction in the short-lived catarina scallop Argopecten ventricosus reared in its natural environment. Comp Biochem Phys A 162(4):421–430. https://doi.org/10.1016/j.cbpa.2012.04.018
Han GD, Zhang S, Marshall DJ, Ke CH, Dong YW (2013) Metabolic energy sensors (AMPK and SIRT1), protein carbonylation and cardiac failure as biomarkers of thermal stress in an intertidal limpet: linking energetic allocation with environmental temperature during aerial emersion. J Exp Biol 216(17):3273–3282. https://doi.org/10.1242/jeb.084269
Hardie DG (2003) Minireview: the AMP-activated protein kinase cascade: the key sensor of cellular energy status. Endocrinology 144(12):5179–5183. https://doi.org/10.1210/en.2003-0982
Hardie DG (2008) AMPK: a key regulator of energy balance in the single cell and the whole organism. Int J Obesity 32(S4):7–12. https://doi.org/10.1038/ijo.2008.116
Hardie DG, Scott JW, Pan DA, Hudson ER (2003) Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett 546(1):113–120. https://doi.org/10.1016/S0014-5793(03)00560-X
Hardie DG, Ross FA, Hawley SA (2012) AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Bio 13(4):251–262. https://doi.org/10.1038/nrm3311
Hawley SA, Davison M, Woods A, Davies SP, Beri RK, Carling D, Hardie DG (1996) Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem 271(44):27879–27887. https://doi.org/10.1074/jbc.271.44.27879
Honkoop P (2003) Physiological costs of reproduction in the Sydney rock oyster Saccostrea glomerata. Oecologia 135(2):176–183. https://doi.org/10.1007/s00442-002-1172-5
Hudson ER, Pan DA, James J, Lucocq JM, Hawley SA, Green KA, Baba O, Terashima T, Hardie DG (2003) A novel domain in AMP-activated protein kinase causes glycogen storage bodies similar to those seen in hereditary cardiac arrhythmias. Curr Boil 13(10):861–866. https://doi.org/10.1016/S0960-9822(03)00249-5
Jost JA, Podolski SM, Frederich M (2012) Enhancing thermal tolerance by eliminating the pejus range: a comparative study with three decapod crustaceans. Mar Ecol Prog Ser 444:263–274. https://doi.org/10.3354/meps09379
Kathyayani SA, Poornima M, Sukumaran S, Nagavel A, Muralidhar M (2019) Effect of ammonia stress on immune variables of Pacific white shrimp Penaeus vannamei under varying levels of pH and susceptibility to white spot syndrome virus. Ecotox Environ Safe 184:109626. https://doi.org/10.1016/j.ecoenv.2019.109626
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360. https://doi.org/10.1038/nmeth.3317
Klopfenstein DV, Zhang L, Pedersen BS, Ramírez F, Vesztrocy AW, Naldi A, Mungall CJ, Yunes JM, Botvinnik O, Weigel M, Dampier W, Dessimoz C, Flick P, Tang H (2018) GOATOOLS: a python library for Gene Ontology analyses. Sci Rep 8:10872. https://doi.org/10.1038/s41598-018-28948-z
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. https://doi.org/10.1038/nmeth.1923
Li YD, Zhou F, Huang JH, Yang L, Jiang S, Yang Q, He JG, Jiang SG (2018) Transcriptome reveals involvement of immune defense, oxidative imbalance, and apoptosis in ammonia-stress response of the black tiger shrimp (Penaeus monodon). Fish Shellfish Immun 83:162–170. https://doi.org/10.1016/j.fsi.2018.09.026
Liang ZX, Liu R, Zhao DP, Wang LL, Sun MZ, Wang MQ, Song LS (2016) Ammonia exposure induces oxidative stress, endoplasmic reticulum stress and apoptosis in hepatopancreas of pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immun 54:523–528. https://doi.org/10.1016/j.fsi.2016.05.009
Lin YC, Chen JC (2001) Acute toxicity of ammonia on Litopenaeus vannamei Boone juveniles at different salinity levels. J Exp Mar Biol Ecol 259(1):109–119. https://doi.org/10.1016/S0022-0981(01)00227-1
Liu CH, Chen JC (2004) Effect of ammonia on the immune response of white shrimp Litopenaeus vannamei and its susceptibility to Vibrio alginolyticus. Fish Shellfish Immun 16:321–334. https://doi.org/10.1016/S1050-4648(03)00113-X
Liu F, Li SH, Yu Y, Sun MZ, Xiang JH, Li FH (2020) Effects of ammonia stress on the hemocytes of the Pacific white shrimp Litopenaeus vannamei. Chemosphere 239:124759. https://doi.org/10.1016/j.chemosphere.2019.124759
Lu X, Kong J, Luan S, Dai P, Meng XH, Cao BX, Luo K (2016) Transcriptome analysis of the hepatopancreas in the pacific white shrimp (Litopenaeus vannamei) under acute ammonia stress. PLoS ONE 11:e0164396. https://doi.org/10.1371/journal.pone.0164396
Ma L, Li ZQ, Bian L, Cai XM, Luo ZX, Zhang YJ, Chen ZM (2016) Identification and comparative study of chemosensory genes related to host selection by legs transcriptome analysis in the tea geometrid Ectropis obliqua. PLoS ONE 11(3):e0149591. https://doi.org/10.1371/journal.pone.0149591
Mai KS, Ai QH, Xu W, Liu Fu ZG (2004) Stress in aquaculture and its prevention with emphasis on nutritional methods. J Ocean U China 34(5):767–774. https://doi.org/10.16441/j.cnki.hdxb.2004.05.13
Martins DA, Rocha F, Martínez-Rodríguez G, Bell G, Morais S, Castanheira F, Bandarra N, Coutinho J, Yúfera M, Conceição LE (2012) Teleost fish larvae adapt to dietary arachidonic acid supply through modulation of the expression of lipid metabolism and stress response genes. Brit J Nutr 108(5):864–874. https://doi.org/10.1017/S0007114511006143
McEwen BS, Wingfield JC (2003) The concept of allostasis in biology and biomedicine. Horm Behav 43:2–15. https://doi.org/10.1016/S0018-506X(02)00024-7
Miranda-Filho KC, Pinho GLL, Wasielesky W Jr, Bianchini A (2009) Long-term ammonia toxicity to the pink-shrimp Farfantepenaeus paulensis. Comp Biochem Phys C 150(3):377–382. https://doi.org/10.1016/j.cbpc.2009.06.001
Peng RB, Le KX, Jiang XM, Wang Y, Han QX (2015) Effects of different diets on the growth, survival, and nutritional composition of juvenile cuttlefish, Sepia pharaonis. J World Aquacult Soc 46:650–664. https://doi.org/10.1111/jwas.12235
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290–295. https://doi.org/10.1038/nbt.3122
Polekhina G, Gupta A, Michell BJ, Van Denderen B, Murthy S, Feil SC, Jennings IG, Campbell DJ, Witters LA, Parker MW, Kemp BE, Stapleton D (2003) AMPK β subunit targets metabolic stress sensing to glycogen. Curr Biol 13(10):867–871. https://doi.org/10.1016/S0960-9822(03)00292-6
Racotta IS, Roberto H (2000) Metabolic responses of the white shrimp, Penaeus vannamei, to ambient ammonia. Comp Biochem Phys A 125(4):437–443. https://doi.org/10.1016/S1095-6433(00)00171-9
Romano N, Zeng C (2013) Toxic effects of ammonia, nitrite, and nitrate to decapod crustaceans: a review on factors influencing their toxicity, physiological consequences, and coping mechanisms. Rev Fish Sci 21(1):1–21. https://doi.org/10.1080/10641262.2012.753404
Rome. http://www.fao.org/3/i9540en/I9540EN.pdf Accessed 7 January 2019
Sánchez-Paz A, García-Carreño F, Hernández-López J, Muhlia-Almazán A, Yepiz-Plascencia G (2007) Effect of short-term starvation on hepatopancreas and plasma energy reserves of the Pacific white shrimp (Litopenaeus vannamei). J Exp Mar Biol Ecol 340(2):184–193. https://doi.org/10.1016/j.jembe.2006.09.006
Shan HW, Dong Y, Ma S, Zhou YG, Ma ZY (2018) Effects of dietary supplementation with freeze-dried powder of Ampithoe sp. on the growth performance, energy metabolism, and ammonia-nitrogen tolerance of the Pacific white shrimp. Litopenaeus Vannamei Aquac Res 49:2633–2643. https://doi.org/10.1111/are.13721
Shan HW, Geng ZX, Ma S, Wang T (2019) Comparative study of the key enzymes and biochemical substances involved in the energy metabolism of Pacific white shrimp, Litopenaeus vannamei, with different ammonia-N tolerances. Comp Biochem Phys C 221:73–81. https://doi.org/10.1016/j.cbpc.2019.04.001
Sirirustananun N, Chen JC, Lin YC, Yeh ST, Liou CH, Chen LL, Sim S, Chiew S (2011) Dietary administration of a Gracilaria tenuistipitata extract enhances the immune response and resistance against Vibrio alginolyticus and white spot syndrome virus in the white shrimp Litopenaeus vannamei. Fish Shellfish Immun 31:848–855. https://doi.org/10.1016/j.fsi.2011.07.025
Tatar M, Carey JR (1995) Nutrition mediates reproductive trade-offs with age-specific mortality in the beetle Callosobruchus maculatus. Ecology 76(7):2066–2073. https://doi.org/10.2307/1941681
Wang T, Shan H, Geng Z, Yu P, Ma S (2020a) Dietary supplementation with freeze-dried Ampithoe sp. enhances the ammonia-N tolerance of Litopenaeus vannamei by reducing oxidative stress and endoplasmic reticulum stress and regulating lipid metabolism. Aquacult Rep 16:100264. https://doi.org/10.1016/j.aqrep.2019.100264
Wang ZL, Feng YY, Li J, Zou JX, Fan LF (2020b) Integrative microRNA and mRNA analysis reveals regulation of ER stress in the Pacific white shrimp Litopenaeus vannamei under acute cold stress. Comp Biochem Phys D 33:100645. https://doi.org/10.1016/j.cbd.2019.100645
Wongsasak U, Chaijamrus S, Kumkhong S, Boonanuntanasarn S (2015) Effects of dietary supplementation with β-glucan and synbiotics on immune gene expression and immune parameters under ammonia stress in Pacific white shrimp. Aquaculture 436:179–187. https://doi.org/10.1016/j.aquaculture.2014.10.028
Xiao Y, Chen J, Xie C, Peng T, Liu Y, Wang W (2017) A diet of fructose-enriched Artemia improves the response of juvenile Litopenaeus vannamei shrimp to acute low-salinity challenge. Aquac Res 48(7):3935–3949. https://doi.org/10.1111/are.13220
Xiao J, Li Q, Tu J, Chen X, Chen X, Liu Q, Liu H, Zhou X, Zhao YZ, Wang H (2019) Stress response and tolerance mechanisms of ammonia exposure based on transcriptomics and metabolomics in Litopenaeus vannamei. Ecotox Environ Safe 180:491–500. https://doi.org/10.1016/j.ecoenv.2019.05.029
Xu C, Li E, Xu Z, Wang S, Chen K, Wang X, Li T, Qin J, Chen L (2016) Molecular characterization and expression of AMP-activated protein kinase in response to low-salinity stress in the Pacific white shrimp Litopenaeus vannamei. Comp Biochem Phys B 198:79–90. https://doi.org/10.1016/j.cbpb.2016.04.005
Zhang J, Liu Y, Tian L, Yang H, Liang G, Xu D (2012) Effects of dietary mannan oligosaccharide on growth performance, gut morphology and stress tolerance of juvenile Pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immun 33:1027–1032. https://doi.org/10.1016/j.fsi.2012.05.001
Zhang XJ, Yuan JB, Sun YM, Li SH, Gao Y, Yu Y, Liu CZ, Wang QC, Lv XJ, Zhang XX, Ma KY, Wang XB, Lin WC, Wang L, Zhu XL, Zhang CS, Zhang JQ, Jin SJ, Yu KJ, Kong J, Xu P, Chen J, Zhang HB, Sorgeloos P, Sagi A, Alcivar-Warren A, Liu ZJ, Wang L, Ruan J, Chu KH, Liu B, Li FH, Xiang JH (2019) Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat Commun 10:356. https://doi.org/10.1038/s41467-018-08197-4
Funding
This work is supported by National Key R&D Program of China, no. 2020YFD0900202 and the Key Laboratory of Mariculture of Ministry of Education, Ocean University of China, no. KLM2018010.
Author information
Authors and Affiliations
Contributions
Teng Wang designed and carried out the study, analyzed the data, and wrote the article; Lei Gao carried out the transcriptome analysis and wrote the article; Wenheng Li analyzed the data; Yun Li analyzed the data; Hongwei Shan designed the study, provided guidance during the research study and helped revise the article. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
All animal experiments and animal protocols were conducted in accordance with the guidelines and approval of the respective Animal Research and Ethics Committees of Ocean University of China (Permit Number: 20141201). The field studies did not involve endangered or protected species.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, T., Gao, L., Li, W. et al. Comparative transcriptome analysis reveals a strategy involving dietary manipulation for reducing the mortality of Litopenaeus vannamei exposed to sublethal ammonia through the energy metabolism pathway. Aquacult Int 30, 1139–1154 (2022). https://doi.org/10.1007/s10499-022-00855-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10499-022-00855-1