Abstract
The swimming crab Portunus trituberculatus is an important farmed species in China. Ammonia-N represents a major physiological challenge for crab culture and the hepatopancreas plays a major role in physiological adaptation processes. To better understand the molecular mechanism of the crab in response to ammonia-N stress, we performed a transcriptome analysis in the hepatopancreas of P. trituberculatus challenged with ammonia-N stress (5 mg/L), using the high-throughput sequencing technology. In total, 52,280 unigenes were obtained from the hepatopancreas of P. trituberculatus, with an average length of 678 bp. Functional categorization and pathways showed some differentially expressed genes were involved in amino acid and nucleobase metabolism, energy metabolism, antioxidation, immune response, reproduction, moulting and material transport. The differential expression patterns of eight randomly selected annotated genes were validated by quantitative real-time PCR (qPCR). Results revealed a substantial number of genes modified by ammonia-N stress and a few significant ammonia acclimation pathways, which will serve as an invaluable resource for revealing the molecular basis of physiological adaptation mechanism in P. trituberculatus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recently, the farming of Portunus trituberculatus has prospered and its dependence on fresh trash fish and feed has increased. Also, the ammonization of organic compounds by microorganisms in the water has increased the likelihood of exposure to high concentration of ammonia-N (Ren et al. 2015). Among water deterioration factors, a high concentration of ammonia-N is the most common environmental limiting factor for crustaceans. Ammonia-N is normally present in water in the ionized (NH4+) and unionized (NH3) states. The relationship between the concentrations of NH3 and NH4+ may be estimated by the Henderson–Hasselbalch equation: pH = pK + log [NH3]/[NH4+]. Physiological solutions act as weak bases (pK of 9.3–9.4), which are mainly present in the protonated form, NH4+. However, NH3 has higher lipid solubility, which makes it easier to diffuse through the phospholipid bilayers (Cameron and Heisler 1983). Ammonia-N has been reported to cause a series of physiological reactions in crustaceans, including mainly behavioral reactions (Zimmer and Wood 2017), antioxidant response (Liang et al. 2016; Pinto et al. 2016), immune stress (Yue et al. 2010), ion regulation (Romano and Zeng 2007), and ammonia excretion process (Wang et al. 2003). Many studies have suggested also that a high concentration of ammonia-N can influence physiological response processes, such as moulting, growth and reproduction, and even lead to death (Chen and Kou 1992; Dutra et al. 2016). Therefore, ammonia-N is now being increasingly considered as a main threat to crustaceans. However, there are only a few studies on the transcriptome level under ammonia-N stress.
A high concentration of ammonia-N has been reported to severely damage the hepatopancreas of crustaceans and even induce the apoptosis of hepatopancreatic cells (Liang et al. 2016). In addition to performing the function of the digestive gland, the hepatopancreas of crustaceans is known as a metabolic factory, which is the center of lipid and carbohydrate metabolism. Furthermore, it exerts an important role in ammonia detoxification, including the conversion of ammonia to urea and uric acid through the ornithine-urea cycle (OUC) (Chen and Chen 1997) and purine nucleotide anabolism (Bernasconi and Uglow 2011). Moreover, amino acid metabolism is carried out and glutamine (Gln) is synthesized mainly through the combined action of glutamate dehydrogenase (GDH) and glutamine synthetase (GS) (Pan et al. 2018). Moreover, the hepatopancreas may serve also as a site for the consumption and storage of organic substances to support a variety of important life activities, such as moulting (Gaxiola et al. 2005), vitellogenin synthesis (Tseng et al. 2001) and ovarian maturation (Chen et al. 1998). The hepatopancreas is one of the immune organs that may serve as a major site for the synthesis and secretion of immune molecules, such as antibacterial peptide (AMP) (Ried et al. 1996), beta-1,3-glucan binding protein (LGBP) (Roux et al. 2002), and lectin or lectin-related proteins (Gross et al. 2001). However, the underlying mechanism of the hepatopancreas in crabs on ammonia-N stress is still unclear and many more genes may be involved in this metabolic process, which needs to be explored. Therefore, large-scale identification of functional genes from hepatopancreas tissue is of great significance and is a necessary condition for studying the metabolic mechanism of crabs.
Next-generation sequencing technologies of 454 Life Sciences, Applied Biosystems (SOLiD sequencing), and Illumina companies have been expertly used to explore the transcriptome information in organisms. Compared with traditional sequencing technologies, next-generation sequencing technologies can provide a large amount of sequence data with a wider range and depth by spending less time and cost (Huse et al. 2007). Recently, digital gene expression (DGE) analysis has become an effective and convenient method for monitoring differences in the transcriptomic response of tissues or organs under environmental stress in aquatic crustaceans, such as P. trituberculatus (Lv et al. 2013), Eriocheir sinensis (Li et al. 2013a; Sun et al. 2014), Sinopotamon henanense (Sun et al. 2016), Macrobrachium rosenbergii (Rao et al. 2015), Procambarus clarkii (Shen et al. 2014), Litopenaeus vannamei (Guo et al. 2016) and Fenneropenaeus chinensis (Li et al. 2013b). Lu et al. (2016) performed a comparative transcriptome analysis of the hepatopancreas between controls and an ammonia-treated group of L. vannamei. 136 significantly differentially expressed genes were detected, of which 94 genes were related to the immune response and other genes were related to growth, apoptosis, moulting and osmoregulation. A study by our group has demonstrated that the response to elevated ambient ammonia-N in the gills involved a variety of physiological and metabolic pathways, mainly involving nucleobase metabolism and amino acid metabolism of P. trituberculatus (Ren and Pan 2014). These studies have considerably enriched our knowledge of the genetics and genomics of crustaceans. However, the differences in transcriptomic response in the hepatopancreas of P. trituberculatus exposed to ammonia-N have not been studied.
The typical benthic swimming crab P. trituberculatus (Crustacea: Decapoda: Brachyura) is one of the most popular aquatic products and is widely cultivated in China due to its rapid growth and high protein content (Ren et al. 2015). In the course of aquacultural practices, ammonia-N concentration may accumulate over time due to the excrement from the cultured animals and decomposition of nitrogenous organic compounds. The present study utilized a high-throughput sequencing technology to analyze transcriptome data obtained from the hepatopancreas of P. trituberculatus experimentally exposed to elevated ambient ammonia-N. The purpose of this study was to discover and investigate the complex molecular responses of P. trituberculatus under ammonia-N stress. The sequencing results are helpful for understanding the physiological functions of the hepatopancreas and provide a foundation for further study of P. trituberculatus (Table 1).
Results
Analysis of DGE libraries
DGE analysis was performed on the hepatopancreas of P. trituberculatus in controls (C1, C2, C3) and the 5 mg/L NH4Cl-exposed group (A1, A2, A3) at 48 h. Over 14 Mio. and 15 Mio. raw reads were generated for control and experiment group libraries, respectively. After trimming the reads including ploy-N, adapter and low-quality reads, a total of 29.5 Mio. clean reads (98.61%) were obtained. Among these clean reads, the percentages of sequences that could be mapped to unigenes in the two libraries were 89.10% and 88.96%, respectively, and the error rates of base sequencing were both 0.03% (Table 2). These sequences were used for subsequent analysis.
Differentially expressed genes between control and ammonia-N group
All tag sequences from six libraries (C1, C2, C3 and A1, A2, A3) were mapped to the P. trituberculatus transcriptome library (SRP018007), which contains 70,569 unigenes. Reads per kilobase of exon model per million mapped reads (RPKM) was used to assess the relative gene expression levels. The percentage of genes in different RPKM intervals was shown in Fig. 1. Differential expression genes (DEGs) were identified by the DGE method. In this study, a total of 52,280 high-quality unigenes with an average length of 678 bp were obtained in the control group and treated group and 60 genes were differentially expressed between the two experimental groups. Under ammonia-N stress, 30 of these transcripts were up-regulated and 30 were down-regulated. The top six up- and down-regulated annotated transcripts in ammonia-treated crabs were listed in Table 3. The most up-regulated transcript in ammonia-treated crabs encode a glycoprotein. The most down-regulated transcript in exposed crabs encode a zinc finger protein. Three down-regulated transcripts encode vitellogenin, which is closely related to reproduction.
Gene ontology (GO) enrichment and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis
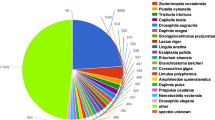
Genes with altered expression covered a variety of physiological metabolic and regulatory processes. In the basis of sequence homology, 14,368 sequences of 52,280 unigenes (27.48%) were classified into 709 subcategories, including 541 biological process terms (76.30%), 100 molecular functions terms (14.10%) and 68 cellular component terms (9.60%). Figure 2 showed that ammonia-responsive genes participated mainly in the nucleobase metabolic process (GO:0019859/GO:0006206/GO:0072527/GO:0009112) and the amino acid metabolic process (GO:0006573/GO:0009081/GO:1901605) according to biological process, oxidoreductase activity (GO:0016620/GO:0016903), methylmalonate-semialdehyde dehydrogenase activity (GO:0004491), and malonate-semialdehyde dehydrogenase activity (GO:0018478) according to molecular functions. For the cellular component terms, no GO term was enriched significantly.
Functional categorization of DEGs in response to environmental ammonia-N in the hepatopancreas of P. trituberculatus-based gene ontology distribution. X-axis represents the number of differential genes of each GO subcategory. GO subcategories are on the Y-axis. “Asterisk” means significantly enriched GO subcategories
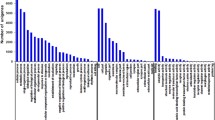
Besides GO analysis, the differentially gene expression caused by elevated ammonia-N affected a range of KEGG pathways. The number of sequences annotated in KEGG pathways ranged from 10 to 2446. The differentially expressed genes were mapped to 45 pathways in the KEGG database and the 20 most enriched KEGG pathways are presented in Fig. 3. The results showed significant enrichment of four KEGG pathways (corrected P value < 0.05). They were beta-alanine metabolism (ko00410, P value = 0.00598), propanoate metabolism (ko00640, P value = 0.00598), valine, leucine and isoleucine degradation (ko00280, P value = 0.00790) and lysosome (ko04142, P value = 0.04050).
Validation of DGE analysis
Eight genes related to the microtubules and vesicle transport, immune system, antioxidant system, reproduction, digestion and moulting were selected to further detect the relative mRNA expression levels by qPCR. These date would verify the gene expression levels identified by DGE. Analysis of the melting curve of qPCR demonstrated that all test genes had a single product. The results showed that the expressions of the MYO7a (involved in microtubules and vesicle transport), CTLD, serine protease (involved in immune response), GPx (involved in antioxidant response), vitellogenin (involved in reproduction), trypsin, CBP B (involved in digestion), ERLP (involved in moulting) using qPCR were consistent with the DGE pattern under ammonia-N exposure (Fig. 4). In theory, the close correlation between qPCR and DGE provides a powerful reference for the quantitative accuracy of DGE method.
Comparison of relative fold change of DGE and qPCR results between the 0 and 5 mg/L NH4Cl groups in the hepatopancreas of P. trituberculatus. The transcript expression levels of the selected genes were each normalized to that of the ribosomal protein L8 (RPL8) gene. MYO7a Myosin-VIIa-like, CTLD C type lectin-containing domain protein, GPx glutathione peroxidase, CBP B carboxypeptidase B, ERLP ecdysteroid-regulated-like protein
Discussion
To better understand the basis of molecular responses and identify key genes and metabolic pathways against ammonia-N stress, we tested the transcriptomic responses of the hepatopancreas in P. trituberculatus under ammonia-N exposure using DGE technology. Previous studies have analyzed the transcriptome from P. trituberculatus using cDNA libraries and Sanger sequencing methods by extracting RNA from gill, hemocytes and eyestalk (Liu et al. 2011; Xu et al. 2010). Recently, transcriptomes in the gills of P. trituberculatus challenged with salinity stress have been analyzed using Illumina Sequencing technology (Lv et al. 2013). 94,511 unigenes were generated by the overall de novo assembly of cDNA sequence data and 1705 genes were differentially expressed under salinity stress. Our previous study generated 58,336 unigenes and detected 69 differentially expressed genes in the gills of the same crab under ammonia-N exposure for 48 h using DGE technology (Ren and Pan 2014). In the present study, DGE technology produced 52,280 unigenes with an average length of 678 bp, which greatly enriched the transcriptome library of crabs and prompted the genome research of crustaceans. However, there are only 60 differentially expressed genes that were detected, of which 30 are up-regulated and 30 are down-regulated, and have been verified by qPCR. Studies have reported that crustaceans can adapt to a relatively high level of ammonia-N after 48 h of exposure (Chen and Nan 1993; Ren and Pan 2014). This may be attributed to the sampling time resulting in limited data acquisition. This suggests that P. trituberculatus has a strong tolerance to ammonia.
For a functional overview of DEGs, an enrichment analysis of GO terms was performed to determine the main biological process that DEGs are involved in. The most significant enriched GO terms that were identified in this experiment include nucleobase and amino acids metabolism. This study was consistent with previous reports, in which the gills in P. trituberculatus were exposed to a relatively high concentration of ammonia-N (Ren and Pan 2014). Also, the hemocytes in L. vannamei were exposed to nitrite-N (Guo et al. 2016). A previous study has suggested that the breakdown of nitrogenous organics was primarily through amino acid catabolism and nucleic acid catabolism (Regnault 1987). Most of the exposed ammonia-N eventually caused the excretion of ammonia (Chen and Cheng 1995). Combined with our results, we speculate that changes in amino acid metabolism associated with nitrogen excretion may be a way to treat endogenous nitrogenous compounds to reduce ammonia toxicity.
In crustaceans, the hepatopancreas is the major metabolic center. KEGG pathway enrichment analysis showed that amino acid (beta-alanine, valine, leucine and isoleucine) and lipid (propanoate) metabolism in hepatopancreas were enriched significantly. In response to ammonia-N exposure, three differentially expressed genes encoding carboxypeptidase B, trypsin and long-chain-fatty-acid–CoA ligase, all of which were involved in the KEGG pathway of protein and lipids digestion, were significantly down-regulated in the hepatopancreas. Similar reports appeared in Farfantepenaeus paulensis post-larvae (Miranda-Filho et al. 2009), and finally showed a significant change in energy metabolism, which is characterized by a decrease in lipid content in the body. This leads to a significant and lengthy decline in growth under high concentrations of ammonia-N. Our study suggested that P. trituberculatus was probably intended to reduce the endogenous ammonia-N level and energy consumption by inhibiting the digestion of food when exposed to ammonia-N. It is well known that metabolism may produce intermediate metabolites, such as free radicals, and eventually cause antioxidant responses. The oxidation–reduction process is always present in the entire life activities, and oxidative stress response is one of the toxic mechanisms of environmental stress factors to aquatic organisms (Valavanidis et al. 2006). Increasing evidence suggests that exposure to ammonia-N can cause oxidative stress in crustaceans, including P. trituberculatus (Ren and Pan 2014), Macrobrachium amazonicum (Pinto et al. 2016), Cherax quadricarinatus (Jiang et al. 2012) and L. vannamei (Liang et al. 2016). It was reported that a high concentration of ammonia may accumulate in the tissues of crustaceans, triggering the release of ROS. Moreover, the concentration of lipid peroxidation in the hepatopancreas of crustaceans will be increased (Cheng et al. 2015; Jia et al. 2017). More importantly, glutathione peroxidase (GPx) could remove lipid peroxides and other organic hydroperoxides by itself. In the present study, molecular functions of GO terms showed that oxidoreductase activity was significantly enriched and the mRNA expression level of GPx was up-regulated at 48 h NH4Cl exposure, with different expression patterns of gills. However, it was consistent with changes in the expression of the hepatopancreas in Exopalaemon carinicauda (Ren et al. 2014) and the liver in Takifugu obscurus (Cheng et al. 2015) under ammonia stress. A similar study involved Dicentrarchus labrax, and unlike other organs (brain, gills, muscle and kidney), hydrogen peroxide (H2O2) and malondialdehyde (MDA) accumulated in the liver when exposed to high environmental levels of ammonia, with a concomitant augmentation in antioxidase activities (Sinha et al. 2015). This may be related to the function and localization of genes in different tissues.
In addition, lysozymes in the hepatopancreas were significantly enriched in the KEGG pathway. Interestingly, two genes related with immune function, including C type lectin containing domain protein and serine protease, were up-regulated in the hepatopancreas when exposed to ammonia-N. Conversely, Ren and Pan (2014) reported that the mRNA expression levels of some immune-related genes were significantly down-regulated under ammonia stress, suggesting that the immune defense capability of crustaceans may be reduced. However, Zhang et al. (2015) reported a significant increase in the immune responses of Megalobrama amblycephala under challenge with high concentrations of ammonia. Moreover, in Carassius auratus and Anoplopoma fimbria, the expression and activity of immune-related factors of the liver were increased under ammonia exposure (Kim et al. 2017; Qi et al. 2017), which were consistent with the results of this study. The functions of the hepatopancreas were related to the synthesis and secretion of immune molecules (Al-Mohanna and Nott 1989), indicating that as the first line of defense with external water, the immune response of the gills was inhibited. This required that the hepatopancreas started the synthesis of immune molecules to maintain homeostasis, resulting in up-regulation of the transcription response under ammonia stress.
Vitellin is the major yolk protein in oviparous animals, which provides nutrition during embryonic development. Vitellogenin, as the precursor of vitellin, may be synthesized in the hepatopancreas and ovary of E. sinensis (Li et al. 2006), Callinectes sapidus (Zmora et al. 2007) and Scylla paramamosain (Jia et al. 2013). Furthermore, a previous study has suggested that the expression of vitellogenin was tissue-specific, including 14 hepatopancreas-specific transcripts and 6 ovary-specific transcripts in P. clarkii (Shen et al. 2014). Here, it was detected that the mRNA expression level of vitellogenin was dramatically decreased in the hepatopancreas during ammonia exposure. Moreover in this database, three sharply down-regulated transcripts encoded the vitellogenin. This indicated that ammonia might destroy the reproductive system and inhibit the development of yolk and embryos in crabs. The results provided a new perspective for studying the reproductive system of P. trituberculatus.
In the present study, moulting-related genes, such as chitinase and ecdysteroid-regulated-like protein, showed significantly reduced expression in response to elevated ammonia-N. Chitinase was the final product of the endocrine cascades of the multiple hormone system used to control crustacean moulting (Zou and Bonvillain 2004). Similar changes in the mRNA expression level of chitinase exposed to ammonia-N have been reported in the hepatopancreas of Penaeus monodon (Zhou et al. 2017). This suggests that ammonia suppressed the moulting of crabs because chitinase was needed to break down the old exoskeleton of crustaceans prior to ecdysis. Also, this was consistent with the findings of the Portunus pelagicus (Liao et al. 2011). In addition, chitinases were apparently the products of ecdysteroid-regulated protein in arthropods. It was reported that the transcription levels of chitinase and ecdysteroid-regulated protein were decreased in the hepatopancreas of L. vannamei under low salinity conditions (Gao et al. 2012). However, studies on the ecdysteroid-regulated protein of the moulting process under ammonia-N stress in crustaceans are scarce. More information is necessary to verify the moulting mechanism of P. trituberculatus when exposed to elevated ambient ammonia-N.
Myosin transports a variety of substances along actin filaments, such as endoplasmic reticulum vesicles, melanosomes, mitochondria, and neuronal secretion particles. Also, myosin is closely related to kinesins and microtubules (Sun et al. 2010). Intriguingly, a substantial increase in Myosin-VIIa-like was detected in this transcriptome library, which can be combined with microtubules and vesicle transport. Moreover, a microtubule-dependent ammonia transport mechanism in the gills of P. trituberculatus (Ren et al. 2015) and Carcinus maenas (Weihrauch et al. 2002) was proposed. Furthermore, this mechanism has been validated in the midgut of Manduca sexta L. (Weihrauch 2006). Yet, more in-depth research is needed to verify the mechanism of ammonia transport in the hepatopancreas of crabs under ammonia-N stress.
Conclusions
In the present study, the next-generation sequencing technique was used to analyze the transcriptome level of key genes in the hepatopancreas of P. trituberculatus under ammonia-N stress. A total of 52,280 unigenes were generated and some differentially expressed genes were identified that were involved in amino acid and nucleobase metabolism. In addition, the physiological responses of P. trituberculatus in ammonia-N stress were closely related to energy metabolism, antioxidation, immune response, reproduction, moulting and material transport. These results revealed that a complex physiological adaptation process existed in P. trituberculatus under elevated ambient ammonia-N condition and provided a foundation for further studies of the molecular response mechanism.
Materials and methods
Experimental animals
The experimental crabs P. trituberculatus (mass 120 ± 6.5 g) were obtained from Nanshan market (Qingdao, China), and acclimated for one week prior to experiments in plastic tanks with aerated sand-filtered seawater at 18 °C ± 0.5 °C and a salinity of 31‰ (pH 8.2). During the acclimation period, two-thirds of the water was renewed twice a day and the crabs were fed with fresh clams Ruditapes philippinarum (10% weight per crab) before the seawater was renewed. Crabs were fasted for two days before the experiment to prevent significant fluctuations in ammonia levels in vivo caused by metabolic ammonia generation after feeding.
Ammonia-N exposure and sample collections
According to the ammonia-N concentration of bottom water in the aquaculture tanks measured by Chen and Zhong (Chen et al. 1988; Zhong et al. 1997), and the ammonia concentration in shrimp and crab polyculture ponds (~ 4 mg/L in Rizhao, China, unpublished), 0.05 mg/L (seawater as control) and 5 mg/L NH4Cl was settled for the experiment. The seawater used in the ammonia-N stress experiment was the same as the seawater used in the acclimation period. During the experiment, the ammonia-N concentrations were measured by hypobromite oxidation (GB17378.4 2007) every 12 h, and the ammonia-N concentrations varied at 0.05 ± 0.03 and 5.23 ± 0.21 mg/L, respectively. Crabs were randomly divided into two groups; each group containing three replicates and each replicate containing six crabs. One group was kept in seawater and used as the control (C1, C2 and C3), and the other group (A1, A2 and A3) was subjected to ammonia-N exposure (5 mg/L NH4Cl) for 48 h. After ammonia exposure, the hepatopancreas of these crabs were sampled randomly from each replicate. Approximately 1–2 g of hepatopancreas was sampled from each crab. The control and ammonia-N treatment group include three replicates in each group for subsequent RNA extraction. The hepatopancreas in each replicate used for RNA extraction was dissected using RNase-treated scissors and forceps and then ground directly in liquid nitrogen. After rapid weighing, 100 mg of tissue powder was placed into 1.5 ml RNase-free tubes and lysed by adding 1 ml RNAiso Plus reagent (TaKaRa, Dalian, China). After sufficient shaking, the lysed sample was centrifuged at 12,000g for 15 min in a frozen centrifuge, and the supernatant was stored in an ultra-low temperature refrigerator at − 80 °C until processed.
Library construction and sequencing
The total RNA of hepatopancreas was extracted using the RNAiso Plus method according to the manufacturer’ s instructions and the DNA removed with RNase-free DNase I (TaKaRa, Dalian, China). RNA was checked for purity (260/280 ratios > 2.0) using a NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). Gel electrophoresis was performed on a 1% agarose gel to verify RNA integrity. Sequencing was performed using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) according to the manufacturer's instructions. mRNA was purified from total RNA using magnetic beads with poly T oligos. Divalent cations were used for fragmentation at elevated temperatures. First- and second-strand cDNA were synthesized. 150–200 bp cDNA fragments and 3 µl USER Enzyme (NEB, USA) were prepared to perform the PCR with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. The AMPure XP system was used for purification of PCR products and the Agilent Bioanalyzer 2100 system was used for evaluating the quality of cDNA libraries. After the clusters were generated, the library preparations were sequenced and 100 bp/50 bp single-end reads were produced by Illumina Hiseq 2000 platform.
Differential gene expression analysis
Clean reads were obtained from the raw data by removing reads containing adapter, ploy-N and low-quality reads. Simultaneously, the Q20, Q30 and GC contents of the clean data were calculated. All downstream analysis was based on the high-quality clean data. The P. trituberculatus transcriptome library (NCBI SRA database: SRP018007) was used to link the expressed sequences to the P. trituberculatus hepatopancreas known gene. RPKM takes into account the effects of sequencing depth and gene length on the reads count, which is a commonly used method to estimate gene expression levels (Mortazavi et al. 2008). DEGs were identified using the DESeq R software package (1.10.1), and the corrected P value < 0.005 and log2 ratio ≥ 1 were set as the threshold for significant differential expression. For pathway enrichment analysis, GO enrichment analysis was performed with the GOseq R package, which corrected for gene length bias. A corrected P value < 0.05 for the GO term was considered to be significantly enriched. Analysis of the enrichment of the pathway based on the KEGG database and evaluation of the statistical enrichment of DEGs in the KEGG pathways were by KOBAS software.
Validation of expression profiles using qPCR
To ensure the accuracy of DGE expression patterns, eight candidate genes were randomly taken for validation by qPCR using the same C1, C2, C3 and A1, A2, A3 group RNA samples as were originally used in DGE sequencing. Total RNA was extracted from the hepatopancreas samples in the control group and the ammonia-N treated group using the Trizol method at 48 h (Invitrogen, USA). According to the manufacturer's protocol, 1 μg of total RNA was reverse transcribed into cDNA in each reaction system using the PrimeScript RT reagent kit with gDNA Eraser (Takara, Dalian, China). Relative mRNA expression levels of candidate genes were measured using specific primers (Table 1) designed by Primer Premier 5.0 software and synthesized by BGI. RpL8 was selected as an internal reference gene after comparing the stability of RpL8 and β-actin (Ren and Pan 2014). Each primer pair was checked by RT-PCR in advance to ensure that it was available for qPCR. This was carried out using the SYBR® PrimeScript™ RT-PCR Kit (TaKaRa, Dalian, China), and experiments were executed with a PikoReal 96 Real-Time PCR System (Thermo Scientific) with a final volume of 10 μl. Each reaction contained 1 μl of cDNA, 5 μl of 2 × SYBR premix Ex taq™ (Takara, Shiga, Japan), 0.2 μl of forward and reverse primers (10 μmol/L), and 3.6 μl of sterile water. The PCR program was set at 95 °C for 30 s and then 40 cycles were performed: 95 °C for 10 s, 55 °C (annealing temperature) for 20 s and 72 °C for 30 s. Relative gene expression levels were assessed using the 2−ΔΔCt method (Pfaffl 2001). PCR efficiency (E) was determined by running standard curves for tenfold serial dilutions of cDNA templates, and calculated according to E = 10[−1/slope]−1 (Rasmussen 2001). For all standard curves, the primer amplification efficiencies of genes were 92–109% and 0.985<R2<0.998. The experimental data were analyzed with one-way analysis of variance (ANOVA) using SPSS version 17.0 (SPSS Inc.) and were finally presented as mean ± standard error (n = 6). P < 0.05 was considered to have a significant difference.
References
Al-Mohanna SY, Nott JA (1989) Functional cytology of the hepatopancreas of Penaeus semisulcatus (Crustacea: Decapoda) during the moult cycle. Mar Biol 101:535–544
Bernasconi CJ, Uglow RF (2011) Purineolytic capacity response of Nephrops norvegicusto prolonged emersion: an ammonia detoxification process. Aquat Biol 11:263–270
Cameron JN, Heisler N (1983) Studies of ammonia in the rainbow trout: physico-chemical parameters, acid-base behaviour and respiratory clearance. J Exp Biol 105:107–125
Chen JC, Chen JM (1997) Arginase specific activity and nitrogenous excretion of Penaeus japonicus exposed to elevated ambient ammonia. Mar Ecol Prog Ser 153:197–202
Chen JC, Liu PC, Lin YT (1988) Super intensive culture of red-tailed shrimp Penaeus penicillatus. J World Aquac Soc 19:127–131
Chen JC, Kou YZ (1992) Effects of ammonia on growth and molting of Penaeus japonicus juveniles. Aquaculture 104:249–260
Chen JC, Nan FH (1993) Effects of ammonia on oxygen consumption and ammonia-N excretion of Penaeus chinensis after prolonged exposure to ammonia. Bull Environ Contam Toxicol 51:122–129
Chen JC, Cheng SY (1995) Hemolymph oxygen content, oxyhemocyanin, protein levels and ammonia excretion in the shrimp Penaeus monodon exposed to ambient nitrite. J Comp Physiol B 164:530–535
Chen Y, Du N, Lai W (1998) Lipid and fatty acid composition of hepatopancreas in different stages of the Chinese mitten crab. Acta Zool Sin 44:420–429
Cheng CH, Yang FF, Ling RZ, Liao SA, Miao YT, Ye CX, Wang AL (2015) Effects of ammonia exposure on apoptosis, oxidative stress and immune response in pufferfish (Takifugu obscurus). Aquat Toxicol 164:61–71
Dutra FM, Forneck SC, Brazão CC, Freire CA, Ballester ELC (2016) Acute toxicity of ammonia to various life stages of the Amazon river prawn, Macrobrachium amazonicum, Heller, 1862. Aquaculture 453:104–109
Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM (2007) Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8:R143
Gao W, Tan B, Mai K, Chi S, Liu H, Dong X, Yang Q (2012) Profiling of differentially expressed genes in hepatopancreas of white shrimp (Litopenaeus vannamei) exposed to long-term low salinity stress. Aquaculture 364:186–191
Gaxiola G, Cuzon G, García T, Taboada G, Brito R, Chimal ME, Paredesa A, Soto L, Rosas C, Van Wormhoudt A (2005) Factorial effects of salinity, dietary carbohydrate and moult cycle on digestive carbohydrases and hexokinases in Litopenaeus vannamei (Boone, 1931). Comp Biochem Physiol A 140:29–39
GB17378.4 (2007) The specification for marine monitoring, part 4: Seawater analysis. Standards Press of China, Beijing, pp 111–113
Gross PS, Bartlett TC, Browdy CL, Chapman RW, Warr GW (2001) Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific white shrimp, Litopenaeus vannamei, and the Atlantic white shrimp, L. setiferus. Dev Comp Immunol 25:565–577
Guo H, Xian JA, Wang AL (2016) Analysis of digital gene expression profiling in hemocytes of white shrimp Litopenaeus vannamei under nitrite stress. Fish Shellfish Immunol 56:1–11
Jia X, Chen Y, Zou Z, Lin P, Wang Y, Zhang Z (2013) Characterization and expression profile of Vitellogenin gene from Scylla paramamosain. Gene 520:119–130
Jia X, Zhang D, Wang F, Dong S (2017) Immune responses of Litopenaeus vannamei to non-ionic ammonia stress: a comparative study on shrimps in freshwater and seawater conditions. Aquacult Res 48:177–188
Jiang Q, Lv L, Jiang G, Minter E, Wang Q, Huang W, Dong S, Yang J (2012) Acute effects of ammonia on antioxidative response and gill Na+/K+ ATPase activity of juvenile Australian red claw crayfish (Cherax quadricarinatus). J Freshw Ecol 27:551–560
Kim JH, Park HJ, Hwang IK, Han JM, Kim DH, Oh CW, Lee JS, Kang JC (2017) Toxic effects of juvenile sablefish, Anoplopoma fimbria by ammonia exposure at different water temperature. Environ Toxicol Pharmacol 54:169–176
Li K, Chen L, Zhou Z, Li E, Zhao X, Guo H (2006) The site of vitellogenin synthesis in Chinese mitten-handed crab Eriocheir sinensis. Comp Biochem Physiol B 143:453–458
Li X, Cui Z, Liu Y, Song C, Shi G (2013a) Transcriptome analysis and discovery of genes involved in immune pathways from hepatopancreas of microbial challenged mitten crab Eriocheir sinensis. PLoS ONE 8:e68233
Li S, Zhang X, Sun Z, Li F, Xiang J (2013b) Transcriptome analysis on Chinese shrimp Fenneropenaeus chinensis during WSSV acute infection. PLoS ONE 8:e58627
Liao YY, Wang HH, Lin ZG (2011) Effect of ammonia and nitrite on vigour, survival rate, moulting rate of the blue swimming crab Portunus pelagicus zoea. Aquacult Int 19:339–350
Liang Z, Liu R, Zhao D, Wang L, Sun M, Wang M, Song L (2016) Ammonia exposure induces oxidative stress, endoplasmic reticulum stress and apoptosis in hepatopancreas of pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immunol 54:523–528
Liu Y, Cui Z, Song C, Wang S, Li Q (2011) Multiple isoforms of immune-related genes from hemocytes and eyestalk cDNA libraries of swimming crab Portunus trituberculatus. Fish Shellfish Immunol 31:29–42
Lu X, Kong J, Luan S, Dai P, Meng X, Cao B, Luo K (2016) Transcriptome analysis of the hepatopancreas in the Pacific white shrimp (Litopenaeus vannamei) under acute ammonia stress. PLoS ONE 11:e0164396
Lv J, Liu P, Wang Y, Gao B, Chen P, Li J (2013) Transcriptome analysis of Portunus trituberculatus in response to salinity stress provides insights into the molecular basis of osmoregulation. PLoS ONE 8:e82155
Miranda-Filho KC, Pinho GLL, Wasielesky W, Bianchini A (2009) Long-term ammonia toxicity to the pink-shrimp Farfantepenaeus paulensis. Comp Biochem Physiol C 150:377–382
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Pan L, Si L, Liu S, Liu M, Wang G (2018) Levels of metabolic enzymes and nitrogenous compounds in the swimming crab Portunus trituberculatus exposed to elevated ambient ammonia-N. J Ocean Univ China 17:957–966
Pfaffl MW (2001) A new mathematical method for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:2002–2007
Pinto MR, Lucena MN, Faleiros RO, Almeida EA, McNamara JC, Leone FA (2016) Effects of ammonia stress in the Amazon river shrimp Macrobrachium amazonicum (Decapoda, Palaemonidae). Aquat Toxicol 170:13–23
Qi XZ, Xue MY, Yang SB, Zha JW, Wang GX, Ling F (2017) Ammonia exposure alters the expression of immune-related and antioxidant enzymes-related genes and the gut microbial community of crucian carp (Carassius auratus). Fish Shellfish Immunol 70:485–492
Rao R, Zhu YB, Alinejad T, Tiruvayipati S, Thong KL, Wang J, Bhassu S (2015) RNA-seq analysis of Macrobrachium rosenbergii hepatopancreas in response to Vibrio parahaemolyticus infection. Gut Pathog 7:6
Rasmussen R (2001) Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara KI (eds) Rapid cycle real-time PCR. Springer, Berlin, pp 21–34
Regnault M (1987) Nitrogen excretion in marine and fresh-water crustacean. Biol Rev 62:1–24
Ren Q, Pan L, Zhao Q, Si L (2015) Ammonia and urea excretion in the swimming crab Portunus trituberculatus exposed to elevated ambient ammonia-N. Comp Biochem Physiol A 187:48–54
Ren Q, Pan L (2014) Digital gene expression analysis in the gills of the swimming crab (Portunus trituberculatus) exposed to elevated ambient ammonia-N. Aquaculture 434:108–114
Ren H, Li J, Li JT, Liang ZX, Liang JP, Ge QQ, Liu P (2014) Effects of acute ammonia stresses on antioxidant enzyme activities and GPx gene expression in Exopalaemon carinicauda. J Agro Environ Sci 33:647–655 (in Chinese with English abstract)
Ried C, Wahl C, Miethke T, Wellnhofer G, Landgraf C, Schneider-Mergener J, Hoess A (1996) High affinity endotoxin-binding and neutralizing peptides based on the crystal structure of recombinant Limulus anti-lipopolysaccharide factor. J Biol Chem 271:28120–28127
Romano N, Zeng C (2007) Acute toxicity of ammonia and its effects on the haemolymph osmolality, ammonia-N, pH and ionic composition of early juvenile mud crabs, Scylla serrata (Forskål). Comp Biochem Physiol A 148:278–285
Roux MM, Pain A, Klimpel KR, Dhar AK (2002) The lipopolysaccharide and β-1, 3-glucan binding protein gene is upregulated in white spot virus-infected shrimp (Penaeus stylirostris). J Virol 76:7140–7149
Shen H, Hu Y, Ma Y, Zhou X, Xu Z, Shui Y, Li C, Xu P, Sun X (2014) In-depth transcriptome analysis of the red swamp crayfish Procambarus clarkii. PLoS ONE 9:e110548
Sinha AK, Zinta G, AbdElgawad H, Asard H, Blust R, De Boeck G (2015) High environmental ammonia elicits differential oxidative stress and antioxidant responses in five different organs of a model estuarine teleost (Dicentrarchus labrax). Comp Biochem Physiol C 174–175:21–31
Sun M, Li YT, Liu Y, Lee SC, Wang L (2016) Transcriptome assembly and expression profiling of molecular responses to cadmium toxicity in hepatopancreas of the freshwater crab Sinopotamon henanense. Sci Rep 6:19405
Sun X, He Y, Hou L, Yang WX (2010) Myosin Va participates in acrosomal formation and nuclear morphogenesis during spermatogenesis of Chinese mitten crab Eriocheir sinensis. PLoS ONE 5:e12738
Sun Y, Zhang Y, Liu Y, Xue S, Geng X, Hao T, Sun J (2014) Changes in the organics metabolism in the hepatopancreas induced by eyestalk ablation of the Chinese mitten crab Eriocheir sinensis determined via transcriptome and DGE analysis. PLoS ONE 9:e95827
Tseng DY, Chen YN, Kou GH, Lo CF, Kuo CM (2001) Hepatopancreas is the extraovarian site of vitellogenin synthesis in black tiger shrimp, Penaeus monodon. Comp Biochem Physiol A 129:909–917
Valavanidis A, Vlahogianni T, Dassenakis M, Scoullos M (2006) Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants. Ecotoxicol Environ Saf 64:178–189
Wang AL, Wang WN, Wang Y, Shang LX, Liu Y, Sun RY (2003) Effect of dietary vitamin C supplementation on the oxygen consumption, ammonia-N excretion and Na+/K+ ATPase of Macrobrachium nipponense exposed to ambient ammonia. Aquaculture 220:833–841
Weihrauch D (2006) Active ammonia absorption inthe midgut of the tobacco hornworm Manduca sexta L.: transport studies and mRNA expression analysis of a Rhesus-like ammonia transporter. Insect Biochem Mol Biol 36:808–821
Weihrauch D, Ziegler A, Siebers D, Towle DW (2002) Active ammonia excretion across the gills of the green shore crab Carcinus maenas: participation of Na+ /K+-ATPase, V-type H+-ATPase and functional microtubules. J Exp Biol 205:2765–2775
Xu Q, Liu Y, Liu R (2010) Expressed sequence tags from cDNA library prepared from gills of the swimming crab, Portunus trituberculatus. J Exp Mar Biol Ecol 394:105–115
Yue F, Pan L, Xie P, Zheng D, Li J (2010) Immune responses and expression of immune-related genes in swimming crab Portunus trituberculatus exposed to elevated ambient ammonia-N stress. Comp Biochem Physiol A 157:246–251
Zhang CN, Li XF, Tian HY, Zhang DD, Jiang GZ, Lu KL, Liu GX, Liu WB (2015) Effects of fructooligosaccharide on immune response, antioxidant capability and HSP70 and HSP90 expressions of blunt snout bream (Megalobrama amblycephala) under high ammonia stress. Fish Physiol Biochem 41:203–217
Zhong S, Chen Y, Ling K, Chen B, Cai Q, Chen M (1997) Studies on variations of contents of NH4+–N, S2− and heterotrophic bacteria in substrate of shrimp ponds and their correlations. J Oceanogr Taiwan Strait 16:449–454
Zhou K, Zhou F, Huang J, Yang Q, Jiang S, Qiu L, Yang L, Zhu C, Jiang S (2017) Characterization and expression analysis of a chitinase gene (PmChi-4) from black tiger shrimp (Penaeus monodon) under pathogen infection and ambient ammonia nitrogen stress. Fish Shellfish Immunol 62:31–40
Zimmer AM, Wood CM (2017) Acute exposure to high environmental ammonia (HEA) triggers the emersion response in the green shore crab. Comp Biochem Physiol A 204:65–75
Zmora N, Trant J, Chan SM, Chung JS (2007) Vitellogenin and its messenger RNA during ovarian development in the female blue crab, Callinectes sapidus: gene expression, synthesis, transport, and cleavage. Biol Reprod 77:138–146
Zou E, Bonvillain R (2004) Chitinase activity in the epidermis of the fiddler crab, Uca pugilator, as an in vivo screen for molt-interfering xenobiotics. Comp Biochem Physiol C 139:225–230
Acknowledgements
The work was supported by the Natural Science Foundation of Shandong Province, China (No. ZR2016CM21). We would like to thank all the laboratory staff for their continuous technical advice, sampling and caring for crabs.
Author information
Authors and Affiliations
Contributions
LS and LP designed the experiments and analyzed the data. HW and XZ performed qPCR experiments. LS conducted ammonia-N stress experiment on crabs and wrote the manuscript with consultation from LP. All authors edited and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that there are no conflict of interest.
Animal and human rights statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Edited by Xin Yu.
Rights and permissions
About this article
Cite this article
Si, L., Pan, L., Wang, H. et al. Transcriptomic response to ammonia-N stress in the hepatopancreas of swimming crab Portunus trituberculatus. Mar Life Sci Technol 2, 135–145 (2020). https://doi.org/10.1007/s42995-020-00033-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42995-020-00033-3