Abstract
Histone modulations have been implicated in various cellular and developmental processes where in Drosophila Mof is involved in acetylation of H4K16. Reduction in the size of larval imaginal discs is observed in the null mutants of mof with increased apoptosis. Deficiency involving Hid, Reaper and Grim [H99] alleviated mof RNAi induced apoptosis in the eye discs. mof RNAi induced apoptosis leads to activation of caspases which is suppressed by over expression of caspase inhibitors like P35 and Diap1clearly depicting the role of caspases in programmed cell death. Also apoptosis induced by knockdown of mof is rescued by JNK mutants of bsk and tak1 indicating the role of JNK in mof RNAi induced apoptosis. The adult eye ablation phenotype produced by ectopic expression of Hid, Rpr and Grim, was restored by over expression of Mof. Accumulation of Mof at the Diap1 promoter 800 bp upstream of the transcription start site in wild type larvae is significantly higher (up to twofolds) compared to mof 1 mutants. This enrichment coincides with modification of histone H4K16Ac indicating an induction of direct transcriptional up regulation of Diap1 by Mof. Based on these results we propose that apoptosis triggered by mof RNAi proceeds through a caspase-dependent and JNK mediated pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mof (males absent on the first) belongs to MYST family of proteins, characterized by highly conserved histone acetyl transferase domains, are involved in a wide range of physiological processes in mammals [1–3]. The MYST family of proteins are of particular interest as its members display diverse roles in various nuclear processes and some of them have been implicated in carcinogenesis [4]. Mof was originally described as one of the essential component of the X chromosome dosage compensation system in Drosophila melanogaster. Mutations in mof are lethal for male fruit flies [5]. Mof is one of the five mammalian MYST family histone acetyl transferases [6] and plays a vital role at multiple points in the cellular DNA damage response and double-strand break [DSB] repair pathways [7, 8]. In Drosophila, Mof is one of the members of the MSL (male sex lethal) complex which is involved in dosage compensation function [9–12]. Recent studies revealed that both Drosophila and mammalian Mof proteins reside in multi-protein complexes and there exists a high level of conservation among the interacting proteins. Both proteins exhibit histone acetyl transferase activity that is specific for histone H4 at K16 [5, 13–16]. Cells expressing the ΔhMof and cells with hMof knock down displayed about three fold higher frequencies of anaphase bridges compared to parental cells. In addition, decreased hMof activity was associated with loss of the cell cycle checkpoint response to DNA double-strand breaks [7]. Human Mof or TIP60 mediated acetylation of p53 at K120 determines whether the cell has to undergo apoptosis or cell cycle arrest [17]. Mutation of the mof gene in mice causes early embryonic lethality and embryos are unable to develop beyond expanded E4.5 blastocyst stage. Also cultures with Mof −/− homozygous mutant morphology die by apoptosis which is characterized by pyknotic nuclei indicating chromatin condensation. The appearance of aberrant chromatin morphology preceded activation of caspase 3 and DNA fragmentation [3]. Flies carrying mof mutation and as well as mof knockdown in SL-2 cells, undergo reduced post-irradiation survival. Depletion of Mof in SL-2 cells also resulted in an elevated frequency of metaphases with chromosomal aberrations, suggesting that the role of MOF in DNA damage response is conserved in Drosophila [18].
Programmed cell death or apoptosis in multi-cellular organisms is necessary both during normal development and tissue homeostasis and as well as to combat irreparable DNA damage [19]. The direct executioners of apoptosis are the caspases or cysteine proteases, which act in a cascade, get activated and can cleave a number of key cellular substrates leading to cell death [20]. In mammals the caspase cascade can be initiated through two major branches, the intrinsic mitochondria pathway and external death receptor pathway [21, 22]. Apoptosis happens throughout fly development. For instance, during embryogenesis, most of midline glial cells are eliminated by apoptosis to achieve a well circuited central neural system [23, 24]. During metamorphosis, steroid hormone ecdysone triggers apoptosis and autophagy to remove the embryonic neural system, larval midgut, salivary glands and some other tissues that are no longer needed during adulthood [25, 26]. In addition, apoptosis occurs during the development of fly compound eye in larva and pupal stages. The fly retina provides a useful model to understand apoptotic events and to elucidate their physiological importance.
Different signals can trigger the regulators of Drosophila apoptosis, namely Hid, Rpr and Grim which serve as activators of caspases. However, keeping caspases in check is vital for normal cell survival. Inhibitor of Apoptosis Proteins (IAPs) act as direct negative regulators of caspases by binding to them and inhibiting their activity [27]. IAPs were discovered in baculoviruses where they share redundant function with the caspase inhibitor protein p35 [28]. In Drosophila, RHG protein encoding genes Hid, Rpr and Grim, located contiguously on the third chromosome are indispensable for apoptosis in fly embryos. Deletion of these genes (RHG) abrogates embryonic apoptosis and cause developmental defects [29–31]. Conversely, over expression of any one of these proteins leads to a reverse effect. The negative regulator of apoptosis, Drosophila Inhibitor of Apoptosis Protein1 (DIAP1), is encoded by the thread (th) locus. The major antagonists of DIAP1 are three pro-apoptotic proteins: Hid, Reaper and Grim. Various apoptosis inducing signals such as ionizing radiation and pathological stimuli can stimulate transcriptional activation of these proteins. Besides, an additional level of post transcriptional regulation is imposed on Hid by the Ras-MAP kinase pathway where activated MAPK brings about an inhibitory phosphorylation on Hid [32]. Every RHG protein binds to DIAP’s BIR (baculoviral IAP repeat) domain via RHG motifs. This prevents DIAP’s inhibition on the cellular caspases, Dronc (initiator caspase-9 ortholog or Drosophila NEDD-2 like caspase) and Drice (Drosophila interleukin converting enzyme, an effector caspase), leading to apoptosis [27, 33]. In contrast to DIAP1, DIAP2 is reported to be dispensable for apoptosis and functions in innate immune response to Gram negative bacterial infection [34]. While evidence about the control of DIAP1 and apoptosis at the level of proteins is ample, not much light has been shed on the regulation of transcription at the th (thread) locus and on the factors involved therein. The ‘guardian of the genome’ p53, an important proapoptotic factor, is structurally conserved in both mammals and flies [35]. While it functions in stress induced apoptosis in both [36], in mammals it also induces cell cycle arrest and senescence [37]. In response to ionizing radiation induced DNA damage, dp53 triggers apoptosis via transcription of rpr [38]. Ectopic over expression of p53 in the Drosophila eye leads to apoptosis [36].
The apoptotic machinery in Drosophila is regulated through three evolutionarily conserved mitogen-activated protein kinase [MAPK] signalling pathways-the extracellular signal-regulated kinase (ERK), the c-Jun N-terminal kinase (JNK) and the p38 pathways [39]. Presence of stress activates cell surface receptors, which transduce these signals to the nucleus via sequential phosphorylation of a series of kinases, headed terminally by MAPKs: ERK, JNK and p38. Activation of the JNK cascade is crucial to several biological processes like proliferation, differentiation and morphogenesis, regeneration, and positive regulation of apoptosis [40, 41]. Genetic and biochemical evidences indicate interaction between proapoptotic proteins Hid, Rpr, Grim and the JNK pathway. Apoptosis in eye discs of Drosophila induced by ectopic expression of Reaper can be partially rescued by increasing the dosage of Bsk (Basket), the Drosophila JNK protein. Also, Rpr mediated degradation of DIAP1 inhibits degradation of DTRAF1, an upstream JNKKK (JNK kinase kinase) and triggers JNK mediated apoptosis [42].
Drosophila Mof plays a major role in maintaining genomic stability during early embryogenesis in addition to DNA damage response [43]. In the present study we have focussed on analysing the role of dmof in apoptotic process during development. Since homozygotes for mof mutation are late larval lethal, we have selected developing larval eye discs and adult eyes of Drosophila as the ideal in vivo tissue to study the interactions of Mof with cell death mediators in the apoptotic signal transduction pathways. Moreover, both larval eye discs and adult eyes do not affect cell viability or fertility and are hence optimal tissues to study functions of genes involved in crucial cellular processes. We found that homozygotes for mof mutation have abnormally small imaginal discs which we attribute to cell autonomous induction of apoptosis rather than cell proliferation. We also show that Mof knockdown in Drosophila eye discs causes phenotypes which resemble JNK gain of function including ectopic apoptosis.
Materials and methods
Drosophila culture and maintenance
mof 1 fly stock was a gift from Prof. John C. Lucchesi and Canton S was used as wild type strain. All crosses were maintained at 25 °C on standard culture media. Common balancer stocks used were obtained from the Bloomington Stock Collection. The genetic markers and phenotypes of each mutation are described in Flybase (http://flystocks.bio.indiana.edu/). Ectopic expression of transgenes was carried out using the UAS–GAL4 system [44]. The glass multiple reporter GMR-GAL4 line [45] was used to drive expression of UAS transgenes in the larval eye discs and adult eyes. All stocks carrying constructs designed to knock down gene expression using the RNAi pathway were obtained from Bloomington Stock Centre. These were generated by the Transgenic RNAi Project (TRiP) (http://www.flyrnai.org/TRiP-HOME.html).
Acridine orange staining
Eye discs from third instar larvae were dissected in PBS and stained with freshly diluted acridine orange solution (1 mg/ml) followed by wash in PBS. The stained discs were mounted in PBS, and viewed immediately under confocal microscope with the Rhodamine Red channel (Excitation wave length: 570 nm and emission wave length of 590 nm) using 10X objective.
Immunostaining of imaginal discs
Eye discs were dissected from well fed 3rd instar larvae, fixed for 20 min in 4 % paraformaldehyde in PBT and then washed in PBT for 10 min. The discs were treated with RNase A (10 mg/ml) at 37 °C for 15 min, washed in PBS, blocked in 2 % animal serum for 1 h and incubated with primary antibody overnight at 4 °C. They were further washed with PBT and PBS (5 min each) and stained with fluorescence conjugated secondary antibody for 4 h. The discs were mounted in Vectashield mounting media (Vector Laboratories, USA) containing DAPI or propidium iodide and viewed under confocal microscope (Olympus FV1000).
Quantitative analysis of stained discs
Stained imaginal discs were processed digitally and fluorescent spots quantified with Image J software (NIH, Bethesda, USA). The region posterior to the morphogenetic furrow in eye disc was selected for analysis. Threshold intensity was set for the spots and region of interest (ROI) was selected. The Area Fraction tab enabled calculation of points with fluorescence intensity greater than the set threshold. Approximately 20 discs for each genotype were analyzed in each experiment.
Diap1-lacZ reporter assay
For 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining, larval eye imaginal discs were fixed in 4 % formaldehyde in PBS for 10 min and then incubated in standard X-Gal staining solution for 4 h at 37 °C.
Total protein isolation and western blot analysis
Nearly 200 eye discs (or 20 larvae) were collected and thoroughly homogenized in lysis buffer (6 % SDS, 1 mM EDTA, 2 mM PMSF, 10 μg/ml Aprotinin, 10 μg/ml Leupeptin, 10 μg/ml Pepstatin). The lysed samples were boiled at 95 °C for 5 min and centrifuged at 12,000 rpm at 4 °C for 10 min. The supernatant was collected and western blot analysis was carried out following standard protocols using MOF (1:300), JNK (1:300), P-JNK (1:300), Diap1 (1:200) and ß-actin (1:500) antibodies (Abcam).
Chromatin immunoprecipitation assay (ChIP assay)
Wild type and mof 1 mutant larvae were grown on standard Drosophila culture media. To detect the amount of association of Mof and H4K16Ac proteins with the Diap1 promoter, chromatin immunoprecipitation was conducted as described previously [46]. The immunoprecipitated sample was amplified by a standard PCR protocol and the ratio of amplified immunoprecipitated DNA to control DNA from three independent experiments was calculated by densitometry. Primers used for amplification are as follows:
Diap1[chip1] | Fwd: 5′-TTGAGGGAAGCCACAATTAGA-3′ Rev: 5′-AATGCGTTCTTTTTGCATCC-3′ |
Diap1[chip2] | Fwd: 5′-ACCAGGCGAAAAGAGTGCTA-3′ Rev: 5′-ATATTTTCGGTGGCGTTCAA-3′ |
Diap1[chip3] | Fwd: 5′-AAGCCCAGAGAGCACTGAAA-3′ Rev: 5′-GCGGTATTGCACAAAATCCT-3′ |
Fwd: Forward primer; Rev: Reverse primer | |
Quantitative PCR (Q-PCR)
Using oligo-d (T) primers and ultra pure RNA (extracted by Trizol method and column purified with Macharey-Nagel kit), first strand cDNA was reverse-transcribed. Relative quantification was performed using SYBR Green Master Mix (Takara Bio) on Real time PCR (Applied Biosystems, 7900HT). Each experiment was repeated thrice independently. Using the 2−ΔΔCT method, fold change in product levels for each sample was determined, as described [47]. 18S rRNA was used for the normalization.
Name | Sequence |
|---|---|
18S rRNA | Fwd: CCTTATGGGACGTGTGCTTT Rev: CCTGCTGCCTTCCTTAGATG |
Mof | Fwd: CTGGGTAGGCTGAGCTATCG Rev: CCAGACGAGGTAATCGGTGT |
Diap1 | Fwd: CCGAGGAACCTGAAACAGAA Rev: GAATCGGCACTGACTTAGCC |
Statistical analysis
Statistical analysis was performed using the graph pad software to evaluate the significant difference between the control and treated samples. The results obtained were expressed as mean ± SD. All the experiments were conducted in triplicates. Statistical significance was assessed using student t test. *** indicates P < 0.001, ** indicates P < 0.01, * indicates P < 0.05.
Results
Mof mutant larvae have smaller imaginal discs than wild type due to increased apoptosis but not reduced cell proliferation
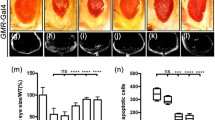
Male mof 1 larvae do not survive past the 3rd instar stage. The mof 1 mutant allele has a Glycine replaced by a Glutamic Acid at position 691 in the acetyl-COA binding motif (i.e. G691E), a change that eliminates its catalytic histone acetyl transferase activity. To understand the cause of lethality in mof 1 mutants, we used fly stocks that carry mof 1 mutation over the balancer FM7GFP. We observed that imaginal discs of mof 1 mutant larvae were significantly smaller in size when compared to wild type discs. The pouch of the wing disc reduced to around 40 % of normal size. We speculated that the small disc phenotype of mof 1 mutants might have resulted either due to reduced cell proliferation or increased apoptosis during the process of development. To assess the role of cell proliferation, we immunostained wing discs from mof 1 mutant larvae using antibody against anti-phospho histone H3 [mitosis marker] that detects mitotic cells. We did not observe any change in the number of mitotic cells in the mutant when compared to wild type [18]. To investigate whether apoptosis is the reason for reduced size of wing discs, mof 1 mutant discs were stained with acridine orange which selectively stains apoptotic cells. In contrast to wild type discs that show little or no apoptosis, mof 1 mutant wing discs displayed robust and wide spread apoptosis (Fig. 1a); clearly depicting apoptosis as the main cause of reduced imaginal disc size in the mof 1 mutants rather than reduced cell proliferation (data not shown). Since mof 1 mutants do not survive beyond the third instar larval stage, in the present study we used the UAS-GAL4 system to knockdown Mof in tissue specific manner using mof RNAi and GMR-GaL4 flies. Eye discs of GMR-GAL4/+; mof RNAi /+ larvae were stained for acridine orange and as expected GMR-GAL4 driven expression of mof RNAi resulted in cell death in eye disc. This was further validated by a genetic rescue experiment by over expressing Mof using EP-Mof flies. The expression of Mof transgene in GMR-mof RNAi background clearly abolished apoptosis caused by knockdown of Mof gene (Fig. 1b).
Small imaginal discs in mof mutants due to increased apoptosis. a Size differences between wing imaginal discs of wild type (Canton S) and mof 1 larvae depicted by acridine orange staining due to increased apoptosis. b Acridine orange stained eye imaginal discs from developing 3rd instar larvae. GMR-GAL4 driven mof RNAi eye discs show increased levels of apoptosis in region posterior to morphogenetic furrow compared to wild type control (CS) which is very much reduced in the presence of Mof transgene. Scale bars represent 50 µm
Drosophila pro-apoptotic genes: Hid, Reaper and Grim are required for mof RNAi induced apoptosis
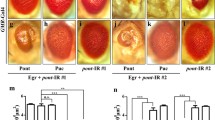
Three regulatory pro-apoptotic proteins Reaper, Hid and Grim (RHG), act as antagonists of Baculovirus, Drosophila and mammalian inhibitor of apoptosis (IAPs) and can predominantly induce apoptosis when ectopically expressed in transgenic animals and cultured insect and mammalian cells [48, 49]. These are a set of closely linked genes encoded by the 75C1region of the Drosophila third chromosome, also known as the H99 region, which when deleted, suppresses apoptosis post γ-irradiation and during embryogenesis [29–31]. To verify the involvement of RHG genes in apoptosis induced by GMR-mof RNAi, we used the deficiency line Df (3L) H99. Interestingly we observed that dosage reduction of reaper, hid and grim genes alleviated GMR-mof RNAi induced apoptosis in the eye discs (Fig. 2a (a–c). Apoptosis in the eye discs induced by GMR-mof RNAi was also considerably reduced in the hid 109 mutant background but the expression of -Hid, Reaper and Grim did not change in the mof mutant at both transcript and protein levels [data not shown]. Ectopic expression of Hid, Rpr and Grim in the adult eye by GMR-GAL4 resulted in ablated apoptotic eye phenotype [29, 50, 51]. To further confirm the genetic interaction of Mof with Hid, Reaper and Grim, we over expressed Mof (P [EP] mof) using GMR-GAL4 in the genetic background of GMR-hid, GMR-reaper and GMR-grim. We observed that over expression of Mof completely recovered the GMR-rpr induced apoptosis but partial recovery was observed in the case of GMR-hid and GMR-grim (Fig. 2b). This is explainable in support with the earlier findings that showed Reaper [and not Hid or Grim] promote the most significant DIAP1 degradation and apoptosis induction [52].
Apoptosis in mof mutants is dependent on pro-apoptotic genes. a Confocal images showing acridine orange staining in eye discs of a GMR-GAL4; mof RNAi /+, b GMR-GAL4; mof RNAi /H99, c GMR-GAL4; mof RNAi /hid 109. b Light microscopic pictures of adult fly eyes of the following genotypes. a CS control, b EP-mof, c GMR-hid, d EP-mof/+; GMR Gal4/+; GMR-hid/+, e GMR-rpr, f EP-mof/+; GMR Gal4/GMR-rpr, g GMR-grim, h EP-mof/+; GMR Gal4/+; GMR-grim/+
Apoptosis caused by down regulation of Mof is suppressed by caspase inhibitors and not p53 mutation
Over expression of baculovirus caspase inhibitor protein p35 and chemical caspase inhibitors inhibit RHG induced ectopic cell death, indicative of their role in caspase mediated cell death [31, 50, 51]. To investigate whether apoptosis induced by GMR-mof RNAi is mediated by caspases we over expressed baculovirus inhibitor of apoptosis P35 [UAS-P35] in the same genetic background. We observed significant reduction of apoptosis in the eye discs, while there was no change in GMR-mof RNAi eye discs in the presence of p53 mutation (data not shown) indicating that apoptosis induced by GMR-mof RNAi is independent of p53. Ark (Apaf-1 related killer) is the Drosophila homolog of mammalian Apaf-1 an essential pro-apoptotic protein and most cell death is blocked in ark mutants [53]. We therefore expressed mof RNAi using GMR-GAL4 in ark RNAi mutant background and observed considerable decrease in apoptosis suggesting the role of ark in mof RNAi induced apoptosis (Figs. 3a (a–c), 2b).
Apoptosis in mof mutants is caspase dependent. a Confocal images showing acridine orange staining in eye discs of a GMR-GAL4; mof RNAi /+, b GMR-GAL4; mof RNAi /UAS-P35, c GMR-GAL4; mof RNAi /Ark RNAi, d GMR-GAL4; mof RNAi /GMR-Diap1, e GMR-GAL4; mof RNAi /diap1 11−3e, f GMR-GAL4; mof RNAi /th 4, g GMR-GAL4; mof RNAi /dronc RNAi, h GMR-GAL4; mof RNAi /drice RNAi. Scale bar represents 50 µm. All images were taken with 10× objective (zoom = 1.8) using Olympus confocal microscope [FV1000]. b Quantification of apoptotic cells. For each genotype 20 discs were analyzed from two independent experiments. Each bar for a genotype represents the mean reading of the fractional fluorescent area over a constant Region of interest (ROI) in the eye discs [Mean ± SEM]
It was earlier reported that mechanism of activation of caspases in Drosophila is mediated by an exclusively important antagonist molecule: the IAPs [33, 54]. This conserved family of inhibitors was first identified in baculovirus [55] by means of their ability to suppress viral-infection. Subsequently homologs were identified in Drosophila melanogaster, C. elegans, mammals, and yeast [56]. To study the role of IAPs, we over expressed Drosophila inhibitor of apoptosis 1 (DIAP1) using GMR-GAL4 in mof RNAi flies. We observed complete rescue of apoptosis confirming that mof mutants induce apoptosis through caspase mediated pathway. We also analysed the effect of loss of function mutants of diap1 [th 4 /+ and diap1 11−3e /+] in the mof RNAi induced apoptosis. As anticipated we observed increased apoptosis in the eye discs of GMR-mof RNAi further validating the antagonistic function of IAPs (Figs. 3a (d–f), 2b).
GMR-mof RNAi induced apoptosis is caused by activation of caspases in the eye discs
Dronc and drICE are Drosophila melanogaster cysteine proteases that function as initiator and effector caspases for programmed cell death at all stages of development. We therefore, co-overexpressed drICE RNAi and dronc RNAi independently with mof RNAi to study their role in cell death. Co-overexpression of mof RNAi along with drice RNAi and dronc RNAi using GMR-GAL4 in two separate experiments substantially reduced apoptosis (Figs. 3a (g–h), 2b). To visualize and detect apoptosis we immunostained eye discs from GMR-mof RNAi larvae using cleaved caspase-3 antibody that recognizes multiple epitopes of Dronc/Dronc-dependent caspases and DrICE. Overexpression of caspases in the GMR region of the eye discs expressing mof RNAi using GMR-GAL4 driver confirmed that caspases mediate mof RNAi induced apoptosis. Discs from GMR-hid that display ectopic apoptosis in eye discs served as positive control (Fig. 4a).
Apoptosis in mof mutants is dependent on JNK pathway. a Immunostained eye discs from GMR-hid [positive control] and GMR > mof RNAi eye discs with anti-caspase3 antibody. Considerable caspase overexpression was observed in the experimental eye disc. Arrows depict fluorescent apoptotic cells. Images taken with a 60× objective (zoom = 1) of Olympus confocal microscope [FV1000]. Scale bar 100 µm. b Acridine orange staining of larval eye discs of GMR-GAL4, GMR-mof RNAi [control], GMsR-mof RNAi in the presence of JNK mutant’s bsk 2, Tak1 RNAi, msn and puc E69. c Western blot analysis of total protein isolated from hs-GAL4; mof RNAi larvae in the presence and absence of heat shock using antibodies against phosphor-JNK and JNK (loading control)
Apoptosis induced by mof RNAi is mediated by JNK pathway
Several recent studies revealed the role of Drosophila c-Jun N-terminal kinase (JNK) signalling in regulation of apoptosis [57–60]. JNK pathway is activated either in apoptotic or in proliferating cells depending on the apoptotic stimuli received [61–63]. Genetic and biochemical analyses have also indicated that c-Jun amino-terminal kinase (JNK) signaling pathway is specifically activated by Transforming growth factor beta (TGF-beta) activated kinase 1 (TAK1 or JNKKK) signaling [64] that regulate cell shape and apoptosis. Ectopic activation of TAK1 also leads to small eye phenotype which is the result of ectopically induced apoptosis. Further Tak1 acts upstream of Bsk in the Drosophila JNK signalling pathway [65]. To determine whether JNK pathway is involved in Mof depletion-induced apoptosis, we blocked JNK activity in the Mof knockdown tissues [GMR-mof RNAi] using JNK mutants. Three lines of evidence have shown that apoptosis is dependent on JNK activity. Firstly apoptosis induced by mof RNAi using GMR-GAL4 in the eye disc was significantly reduced in the presence of bsk 2 (JNK) and TAK1 RNAi (JNKKK) mutants. Second apoptosis in Mof knock down eye discs was elevated when Msn (MAPKKKK) was over expressed using UAS-Msn flies and in the genetic background of Puc E69 [inhibitor of JNK] (Fig. 4b). Taken together, results demonstrated that Mof knockdown requires JNK signalling pathway to induce cell death in Drosophila. Thirdly, we also addressed JNK activity directly by analysing the expression of phosphorylated JNK, which is an activated form of the, protein in wild type versus the hs-mof RNAi. A drastic increase in the expression of P-JNK in hs-mof RNAi larvae upon heat shock was observed when compared to wild type further confirming the involvement of JNK pathway in Mof induced apoptosis (Fig. 4c).
Drosophila Mof activates Diap1
The JNK signaling has been reported to be controlled by either Drosophila inhibitor of apoptosis (DIAP1) or the initiator caspase Dronc [66–68]. Drastic reduction of DIAP1 transcript was observed in the mof 1 mutation, but surprisingly, dronc (Drosophila caspase-9 homolog), which is regulated directly by DIAP1, did not show any change in transcript levels. Similar results were obtained with drice transcripts posing a probability that caspase activation immediately precedes cell death (Fig. 5a). Similar to the mRNA levels, DIAP1 protein was also decreased in mof 1 mutant larvae (Fig. 5b). Over expression of Mof in the larval eye discs using GMR-Gal4 resulted in increased expression of DIAP1 which was further confirmed by immunostaining of the eye discs with anti-DIAP1antibody. This pattern of DIAP1 staining is similar to earlier published reports [55]. Also Lac Z staining of eye discs of GMR-Gal4 driven EP-Mof/diap1-LacZ flies has shown a similar expression pattern of diap1 (Fig. 5c). All the above studies clearly confirmed that Mof activates Diap1.
Mof interacts genetically with Diap1. a RT-PCR analysis of total RNA extracted from Canton S and mof 1 larvae using primers against genes Diap1, Dronc and Drice. 18S rRNA is used as loading control. b Western blot analysis of total protein isolated from Canton S and mof 1 larvae using antibodies against Mof and Diap1. β-actin is used as loading control. c Immunostaining of eye discs from larvae over expressing of Mof (GMR > EP-mof ] with anti-DIAP1 antibody). Red colour indicates propidium iodide staining, green indicates anti DIAP1 stain and merge is the superimposition of PI stained eye discs with the green to show the exact location of the expression of DIAP1. Lac Z staining of larval eye discs of genotype GMR > EP-mof; diap1-lacz. Scale bar 50 µm (Color figure online)
Mof regulates the transcription of diap1 by binding to its promoter sequence
Since Mof interacts genetically with Diap1, the next question was to study the mechanism of regulation of Diap1. Till date, transcriptional regulation of Diap1 has not been explored except reports of positive regulator STAT92E binding the Diap1promoter at a distance of 3.3 kb upstream from the transcription start site (TSS) [69]. Berkeley Genome Project Promoter Prediction Tool has been used to predict around 8 possible promoter stretches, lying between the +1TSS and the STAT site-bounded 1.2 kb region. PCR primers of 100–250 bp amplicons spanning the promoter stretch were designed for carrying out ChIP assay (Fig. 6a, b). ChIP assay with antibody against Mof in wild type Canton S third instar larvae revealed enrichment of Mof at all the amplicons. Similar levels of enrichment at all the stretches were obtained using anti-H4K16Ac antibody in wild type larvae confirming that histone modifications are the result of binding of Mof. In parallel, mof 1 mutant larvae as negative control did not show any binding at the Diap1 promoter (Fig. 7a–c). These results clearly established the role of Mof as a transcriptional activator of Diap1 (Fig. 8).
Promoter sequence of Diap1 gene. a diap1 promoter as predicted by BDGP promoter prediction tool [http://tools.genome.duke.edu/generegulation/McPromoter/]. b Primer positions on the DIAP1 promoter, upstream of the +1 transcription start site (TSS). F forward primer, R reverse primer. The previously reported STAT binding sites (SBS) lie further upstream
Chromatin immunoprecipitation assay to detect Mof binding at the diap1 promoter. RT-PCR on ChIP DNA from CS and mof 1 mutant larvae has been carried out. Lane 1 Input (5 % of cross-linked chromatin), Lane 2 ChIP DNA, Lane 3 No antibody control. a, b Quantitative PCR of DNA immunoprecipitated from wild-type CS and mof 1 mutant larval discs Antibodies used: Mof and antiH4K16Ac. c Each bar represents the mean fold enrichment from two independent experiments (with error bars denoting + SEM). Enrichment was calculated as the fold difference (\(2^{{ - {\varDelta \varDelta }C_{t} }}\)) between DCt oftarget sample (Ct value of ChIP sample normalized to input sample) and DCt of negative control (calibrator sample). No antibody control was chosen as the negative control since no deletion mutant of mof is available. P value <0.05 (two-tailed student’s t test)
Hypothesis depicting the role of Drosophila Mof in Apoptosis. a In wild type Mof binds to Diap1 promoter thereby acetylating at H4K16 leading to increased expression of Diap1 there by inducing apoptosis in JNK mediated pathway. b (A). In the wild type Mof, expression of Diap1 is more and it inhibits apoptosis by regulating hid, reaper, Grim via BIR domain of Diap1. Hence apoptosis does not occur. B In mof RNAi flies the production Diap1 is less since H4K16Ac is less at Diap1 promoter. Higher expression of Hid, Reaper and Grim (RHG) degrades diap1 protein. This leads to more production of Dronc and Drice and ultimately apoptosis via Tak1-Bsk signalling molecules that play key role in JNK signalling pathway. Thus there is direct regulation between Diap1 mediated apoptotic signalling through JNK pathway. In simple Mof controls Diap1 and Diap1 modulates apoptotic signalling via Tak1 and Bsk protein that play pivotal role in JNK signalling. C Mof mutation or mof RNAi (having reduced Diap1 protein expression) in the background of JNK mutants tak1 and bsk mitigates apoptosis
Discussion
In this study to address the novel function of Drosophila Mof in the regulation of apoptosis, we carried out in vivo experiments involving both loss and gain of function experiments. We attributed the reasons for the reduced size of larval imaginal discs upon Mof knockdown to increased apoptosis rather than reduced cell proliferation. The fact that apoptosis induced by depletion of Mof occurs through an activated JNK pathway raises questions about the possibility of Mof interaction with downstream regulators of JNK. Mof, a MYST histone acetyltransferase is an essential component of the Drosophila Dosage Compensation Complex (DCC) and shares orthologs from yeast to humans [70]. Until recent times, only few studies have been carried out to explore the possible functional roles of Drosophila homolog of human Mof. In mammals Mof is involved in embryonic development and maintenance of normal epigenetic signature during early stages of development [71, 72]. Depletion of Mof results in decreased levels of H4K16Ac in both mammals and Drosophila with associated alterations in gene transcription but with minimum affect on transcription of DNA repair genes [8, 73].
We have demonstrated for the first time that depletion of Mof in Drosophila induces apoptosis leading to reduced size of the larval imaginal discs. Since mof mutants do not survive beyond the larval stage, mof RNAi flies have been used to study the depletion of Mof in larval imaginal discs. Depletion of Mof in GMR-mof RNAi flies in the genetic background of Df (3L) H99 (deletion of Reaper, Hid and Grim) rescued apoptosis in the GMR region of the eye disc indicating the role of RHG genes in Mof induced apoptosis. The evolutionary conserved pathway of cell death is intrinsic and performed by an army of caspases, Apaf-1 and Bcl-2 members in most of the organisms [74, 75]. The executioners of apoptosis in flies, as well as in C. elegans and humans belong to an evolutionary conserved family of highly selective proteases or caspases [76]. The role of caspases in apoptosis induced by GMR-mof RNAi was elucidated by co-expression of caspase inhibitors P35 and Diap1. Similar to mammalian p53, over expression of Drosophila p53 leads to apoptosis but does not cause G1 arrest. Moreover, p53 mutants do not affect X ray-induced cell cycle arrest [77]. Apoptosis induced by GMR-mof RNAi remained unchanged in the presence of p53 mutation clearly demonstrating that apoptosis induced by GMR-mof RNAi is independent of p53 in Drosophila. Apoptosis in GMR-mof RNAi discs was significantly reduced in the presence of drICE RNAi and dronc RNAi elucidating caspase mediated cell death in the eye discs of GMR-mof RNAi larvae. Loss of function mutations of diap1 are embryonic lethal while ectopic expression can inhibit apoptosis [51]. The primary mode of regulation occurs through its targeted ubiquitination and proteosomal degradation of initiator caspase DrONC [78]. Ectopic expression of pro DRONC in the Drosophila eye generates a severe eye ablation phenotype, which was completely rescued by co-expression of Diap1. However such reversion fails when the ablation phenotype was induced by prodomain lacking DRONC, thus confirming the specific interaction between Diap1 and prodomain of Dronc. We observed drastic increase in the expression of diap1 upon immunostaining of GMR > Mof eye discs with Diap1 antibody and diap1-LacZ staining. A drastic decrease in Diap1 transcripts and protein was observed in the mof 1 mutants indicating that Mof regulates the expression of Diap1.
In wild type, Mof as Histone acetyl transferase enhanced Diap1 promoter acetylation leading to its activation but was highly decreased in the case of mof mutation. Hence increased expression of Diap1 leads to decreased apoptosis. In General RHG proteins such as Hid, Reaper and Grim degrade the Diap1 by interacting with BIR domain of Diap1 protein in the background of mof mutation. But in case of wild-type flies as a genomic stability maintainer, Mof increases the Diap1 expression levels and decreases the pro-apoptotic proteins such as Hid, Reaper, Grim and caspases Drice and Dronc. Interestingly JNK signalling has been reported to be controlled by either Drosophila DIAP1 or the initiator caspase Dronc [66–68].
It has been reported earlier that extracellular death-inducing signals might be transduced via JNK pathway in Drosophila [79]. Cell death induced by GMR > mof RNAi is accompanied by activation of JNK pathway in the eye discs. Cell death induced by GMR > mof RNAi is rescued in the presence of mutants of the JNK pathway or its negative regulator. Our genetic assays point towards the involvement of Tak1, Hep and Bsk as crucial JNK components in Mof-induced apoptosis. Increased phosphorylation of JNK molecules in GMR > mof RNAi tissue definitively points towards the involvement of JNK signalling in apoptosis. Thus our results clearly indicate that Mof regulates Diap1 and Diap1 in turn is regulated by JNK signalling (i.e. particularly the key components of JNK pathway such as Tak1 and Bsk). This provides a different dimension to that of existing one that Diap1 regulates JNK signalling [66–68]. We suspect that a feed-back or interlinking mechanism exists between Diap1 and JNK signalling during apoptosis.
We show that rough eye phenotype induced by GMR-Rpr in the adult eye was completely rescued by over expression of Mof and partially in the case of GMR-hid and GMR-grim. These results clearly elucidated the genetic interaction between pro-apoptotic genes and Mof. It has also been well documented that certain interdependent regulatory loop exist between Diap1 proteins with RHG components ubiquitinating and destroying Diap1 and also Diap1 in maintaining RHG stability to a certain extent [79]. Drosophila melanogaster tumour-necrosis factor receptor-associated factor 1 (DTRAF1) has been identified as a dominant suppressor of Reaper-induced cell death. It was also shown that Reaper modulates the JNK pathway through Drosophila DIAP1, which negatively regulates DTRAF1 by proteasome-mediated degradation [42]. Moreover, Diap1 also possesses the ability to bring about in vitro ubiquitination of the caspase Dronc [80, 81]. We have demonstrated using chromatin immunoprecipitation assays that Mof binds to the promoter regions of Diap1 and thereby regulate its transcriptional activity. Hid, Grim and Reaper are the essential death–inducing genes (required for the destruction of larval tissues), functioning through destruction of Diap1 during development. However, no true antagonist proteins have been identified till date. As a chromatin modifier, Mof promotes transcriptional activation of DIAP1 and act to antagonize unnatural cell death. Functioning against apoptosis promoting proteins-Hid, Reaper and Grim, Mof helps in maintaining optimal cellular levels of DIAP1 especially in the cell’s decision to undergo apoptosis. Our studies led us to hypothesise through genetic and biochemical evidences that Mof is one of those candidate proteins which actually induce Diap1 transcription. It probably helps to maintain a threshold level of Diap1 in the cell and overproduce it upon receiving appropriate signals. We believe, however that this may not be the sole method of regulation of Diap1 by Mof. Further studies need to be carried to determine if Mof can inhibit Diap1 by other mechanisms, such as inhibiting its ubiquitination.
References
Avvakumov N, Cote J (2007) The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 26:5395–5407
Rea S, Xouri G, Akhtar A (2007) Males absent on the first (MOF): from flies to humans. Oncogene 26:5385–5394
Thomas T, Voss AK (2007) The diverse biological roles of MYST histone acetyltransferase family proteins. Cell Cycle 6:696–704
Yang XJ (2004) The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res 32:959–976
Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC (1997) mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J 16:2054–2060
Neal KC, Pannuti A, Smith ER, Lucchesi JC (2000) A new human member of the MYST family of histone acetyl transferases with high sequence similarity to Drosophila MOF. Biochim Biophys Acta 1490:170–174
Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK (2005) Involvement of human MOF in ATM function. Mol Cell Biol 25:5292–5305
Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, Cote Pandita TK (2010) MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol 30:3582–3595
Lucchesi JC, Kelly WG, Panning B (2005) Chromatin remodeling in dosage compensation. Annu Rev Genet 39:615–651
Mendjan S, Akhtar A (2007) The right dose for every sex. Chromosoma 116:95–106
Rea S, Akhtar A (2006) MSL proteins and the regulation of gene expression. Curr Top Microbiol Immunol 310:117–140
Straub T, Becker PB (2007) Dosage compensation: the beginning and end of generalization. Nat Rev Genet 8:47–57
Akhtar A, Zink D, Becker PB (2000) Chromodomains are protein-RNA interaction modules. Nature 407:405–409
Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC (2000) The drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol 20:312–318
Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC (2005) A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol 25:9175–9188
Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A (2005) hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol 25:6798–6810
Tang Y, Luo J, Zhang W, Gu W (2006) Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 24:827–839
Bhadra MP, Horikoshi N, Pushpavallipvalli SN, Sarkar A, Bag I, Krishnan A, Lucchesi JC, Kumar R, Yang Q, Pandita RK, Singh M, Bhadra U, Eissenberg JC, Pandita TK (2012) The role of MOF in the ionizing radiation response is conserved in Drosophila melanogaster. Chromosoma 121:79–90
Jacobson MD, Weil M, Raff MC (1997) Programmed cell death in animal development. Cell 88:347–354
Hengartner MO (2000) The biochemistry of apoptosis. Nature 407:770–776
Danial NN, Korsmeyer SJ (2004) Cell death: critical control points. Cell 116(2):205–219
Salvesen GS, Abrams JM (2004) Caspase activation: stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene 23(16):2774–2784
Sonnenfeld MJ, Jacobs JR (1995) Apoptosis of the midline glia during Drosophila embryogenesis—a correlation with axon contact. Development 121(2):569–578
Zhou L, Hashimi H, Schwartz LM, Nambu JR (1995) Programmed cell-death in the Drosophila central-nervous-system midline. Curr Biol 5(7):784–790
Jiang CG, Baehrecke EH, Thummel CS (1997) Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124(22):4673–4683
Baehrecke EH (2003) Autophagic programmed cell death in Drosophila. Cell Death Differ 10(9):940–945
Goyal L, McCall K, Agapite J, Hartwieg E, Steller H (2000) Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J 19:589–597
Clem RJ, Miller LK (1994) Control of programmed cell death by the baculovirus genes p35 and iap. Mol Cell Biol 14:5212–5222
Grether ME, Abrams JM, Agapite J, White K, Steller H (1995) The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev 9:1694–1708
White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H (1994) Genetic control of programmed cell death in Drosophila. Science 264:677–683
Chen P, Nordstrom W, Gish B, Abrams JM (1996) Grim, a novel cell death gene in Drosophila. Genes Dev 10:1773–1782
Bergmann A, Agapite J, McCall K, Steller H (1998) The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 95:331–341
Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA (1999) The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98:453–463
Huh JR, Foe I, Muro I, Chen CH, Seol JH, Yoo SJ, Guo M, Park JM, Hay BA (2007) The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J Biol Chem 282:2056–2068
Sogame N, Kim M, Abrams JM (2003) Drosophila p53 preserves genomic stability by regulating cell death. Proc Natl Acad Sci USA 100:4696–4701
Fan Y, Lee TV, Xu D, Chen Z, Lamblin AF, Steller H, Bergmann A (2010) Dual roles of Drosophila p53 in cell death and cell differentiation. Cell Death Differ 17:912–921
Zilfou JT, Lowe SW (2009) Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol 1:a001883
Brodsky MH, Sekelsky JJ, Tsang G, Hawley RS, Rubi GM (2000) mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev 14:666–678
Schaeffer HJ, Weber MJ (1999) Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol Cell Biol 19:2435–2444
Davis RJ (2000) Signal transduction by the JNK group of MAP kinases. Cell 103:239–252
Weston CR, Davis RJ (2002) The JNK signal transduction pathway. Curr Opin Genet Dev 12:14–21
Kuranaga E, Kanuka H, Igaki T, Sawamoto K, Ichijo H, Okano H, Miura M (2002) Reaper-mediated inhibition of DIAP1-induced DTRAF1 degradation results in activation of JNK in Drosophila. Nat Cell Biol 4:705–710
Pushpavalli SN, Sarkar A, Bag I, Hunt CR, Ramaiah MJ, Pandita TK, Bhadra U, Pal-Bhadra M (2014) Argonaute-1 functions as a mitotic regulator by controlling Cyclin B during Drosophila early embryogenesis. FASEB J 28:655–666
Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415
Kohler G, Milstein C (1975) Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256:495–497
Cavalli G, Paro R (1999) Epigenetic inheritance of active chromatin after removal of the main transactivator. Science 286:955–958
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25:402–408
Vucic D, Kaiser WJ, Harvey AJ, Miller LK (1997) Inhibition of reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs). Proc Natl Acad Sci USA 94:10183–10188
McCarthy JV, Dixit VM (1998) Apoptosis induced by Drosophila reaper and grim in a human system. Attenuation by inhibitor of apoptosis proteins (cIAPs). J Biol Chem 273:24009–24015
Bump NJ, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P et al (1995) Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science 269:1885–1888
Hay BA, Wassarman DA, Rubin GM (1995) Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83:1253–1262
Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H (2002) Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol 4:432–438
Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14:1902–1910
Lisi S, Mazzon I, White K (2000) Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics 154:669–678
LaCasse EC, Baird S, Korneluk RG, MacKenzie AE (1998) The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene 17:3247–3259
Uren AG, Coulson EJ, Vau DL (1998) Conservation of baculovirus inhibitor of apoptosis repeat proteins (BIRPs) in viruses, nematodes, vertebrates and yeasts. Trends Biochem Sci 23:159–162
Glavic A, Molnar C, Cotoras D, Celis JF (2009) Drosophila Axud1 is involved in the control of proliferation and displays pro-apoptotic activity. Mech Dev 126:184–197
Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T, Miura M (2002) Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J 21:3009–3018
Wu C, Chen C, Dai J, Zhang F, Chen Y, Li W, Pastor-Pareja JC, Xue L (2015) Toll pathway modulates TNF-induced JNK-dependent cell death in Drosophila. Open Biol 5(7):140171. doi:10.1098/rsob.140171
Yang SA, Su MT (2011) Excessive Dpp signalling induces cardial apoptosis through dTAK1 and dJNK during late embryogenesis of Drosophila. J Biomed Sci 18:85. doi:10.1186/1423-0127-18-85
Ryoo HD, Gorenc T, Steller H (2004) Apoptotic cells can induce compensatory cellproliferation through the JNK and the Wingless signalling pathways. Dev Cell 7:491–501
Perez-Garijo A, Shlevkov E, Morata G (2009) The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development 136:1169–1177
Bergantinos C, Corominas M, Serras F (2010) Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development 137:1169–1179
Takatsu Y, Nakamura M, Stapleton M, Danos MC, Matsumoto K, O’Connor MB, Shibuya H, Ueno N (2000) TAK1 participates in c-Jun N-terminal kinase signaling during Drosophila development. Mol Cell Biol 20(9):3015–3026
Gregory CD (2013) Death in the nervous system: JNK signaling in junk clearance. Cell Death Differ 20:1125–1127
Huh JR, Guo M, Hay BA (2004) Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of apical cell death caspase Dronc in a non-apoptotic role. Curr Biol 14:1262–1266
Wells BS, Yoshida E, Johnston A (2006) Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr Biol 16:1606–1615
Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M (2006) DRONC coordinates cell death and compensatory proliferation. Mol Cell Biol 26:7258–7268
Betz A, Ryoo HD, Steller H, Darnell JE Jr (2008) STAT92E is a positive regulator of Drosophila inhibitor of apoptosis 1 (DIAP/1) and protects against radiation-induced apoptosis. Proc Natl Acad Sci USA 105:13805–13810
Conrad T, Akhtar A (2011) Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet 13:123–134
Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK, Ludwig T, Pandita TK (2008) The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol 28:397–409
Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S (2009) Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138:1122–1136
Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, Akhtar A (2008) Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell 133:813–828
Vernooy SY, Copeland J, Ghaboosi N, Griffin EE, Yoo SJ, Hay BA (2000) Cell death regulation in Drosophila: conservation of mechanism and unique insights. J Cell Biol 150:F69–F76
Shi Y (2001) A structural view of mitochondria-mediated apoptosis. Nat Struct Biol 8:394–401
Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J (1996) Human ICE/CED-3 protease nomenclature. Cell 87:171
Ollmann M, Young LM, DiComo CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, Duyk G, Friedman L, Prives C, Kopczynski C (2000) Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101:91–101
Palaga T, Osborne B (2002) The 3D’s of apoptosis: death, degradation and DIAPs. Nat Cell Biol 4:E149–E151
Bergmann A, Yang AY, Srivastava M (2003) Regulators of IAP function: coming to grips with the grim reaper. Curr Opin Cell Biol 15:717–724
Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H, Meier P (2002) The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol 4:445–450
Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, Shi Y (2003) Molecular mechanism of Reaper–Grim–Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol 10:892–898
Acknowledgments
This work has been supported by Department of Biotechnology to MPB [GAP 0362] and UB. The authors thank Prof. John C. Lucchesi for fly stocks. AS, GKR thank UGC for their fellowship and IB thank CSIR for her postdoctoral fellowship. All the authors thank P. Devender for culturing and maintaining the Drosophila stocks. Our thanks to Hemalatha for help in formatting the work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Sreerangam N. C. V. L. Pushpavalli, Arpita Sarkar and M. Janaki Ramaiah have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Pushpavalli, S.N.C.V.L., Sarkar, A., Ramaiah, M.J. et al. Drosophila MOF regulates DIAP1 and induces apoptosis in a JNK dependent pathway. Apoptosis 21, 269–282 (2016). https://doi.org/10.1007/s10495-015-1206-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-015-1206-1