Abstract
Impaired mitochondrial integrity and function are key features of intrinsic death pathways in neuronal cells. Therefore, key regulators of intrinsic death pathways acting upstream of mitochondria are potential targets for therapeutic approaches of neuroprotection. The tumor suppressor p53 is a well-established regulator of cellular responses towards different kinds of lethal stress, including oxidative stress. Recent reports suggested that p53 may affect mitochondrial integrity and function through both, transcriptional activation of mitochondria-targeted pro-death proteins and direct effects at the mitochondrial membrane. In the present study, we compared the effects of pharmacological inhibition of p53 by pifithrin-α with those of selective p53 gene silencing by RNA interference. Using MTT assay and real-time cell impedance measurements we confirmed the protective effect of both strategies against glutamate-induced oxidative stress in immortalized mouse hippocampal HT-22 neurons. Further, we observed full restoration of mitochondrial membrane potential and inhibition of glutamate-induced mitochondrial fragmentation by pifithrin-α which was, in contrast, not achieved by p53 gene silencing. Downregulation of p53 by siRNA decreased p53 transcriptional activity and reduced expression levels of p21 mRNA, while pifithrin-α did not affect these endpoints. These results suggest a neuroprotective effect of pifithrin-α which occurred at the level of mitochondria and independently of p53 inhibition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Degeneration and death of neurons are the major underlying causes for impaired brain functions in many neurological disorders such as Alzheimer’s disease (AD), Parkinson’s disease (PD) and stroke [1]. Although these diseases show significant differences in their pathogenesis and symptoms, they all share common mechanistic features such as the involvement of oxidative stress which can trigger neurodegeneration [2]. Further, there is increasing evidence for a pivotal role of increasing mitochondrial impairments during processes of neurodegeneration and aging [3]. In fact, the loss of mitochondrial integrity and function is the hallmark of intrinsic pathways of programmed cell death (PCD). The translocation of the proapoptotic protein Bid to mitochondria [4] as well as the detrimental generation of reactive oxygen species (ROS) results in the permeabilization of the mitochondrial membrane [5]. Once this point of mitochondrial demise has been reached, the death of neuronal cells is inevitable [6]. Thus, the protection of mitochondria is considered to be a potential therapeutic target in neurological disorders. For example, restoration of mitochondrial function improved performance in a transgenic mouse model of AD [7], and even in humans beneficial effects of “mitoprotection” against progression of neurodegenerative processes were observed [8].
The tumor suppressor protein p53 is regarded as the guardian of the genome. Hence, it plays a crucial role in sensing DNA damage, cell cycle regulation and cellular response to oxidative stress affecting DNA integrity, also in neurons. Dysfunction of p53 contributes to neuronal impairment in AD and PD [9]. Depending on the nature and the extent of cellular stress and associated DNA damage, p53 can either mediate DNA repair via transcriptional induction of p21 [10] or cell death by induction of e.g. the Bcl-2-family protein BAX and the BH3-only proteins Bid, PUMA and NOXA [1, 11, 12]. On the one hand, a loss of function in the p53 gene is often associated with an enhanced responsiveness to cancer [13], but on the other hand, p53 knockout or pharmacological inhibition of p53 was shown to be beneficial in cases of neuronal damage [14–16]. These findings underline the importance of a well controlled p53 expression in terms of disease. Besides transcriptional activity, p53 can also directly act at the level of mitochondria to induce PCD [17–19].
As the induction of apoptosis is one of the most important functions of p53, its pharmacological inhibition has been studied for a long time as a potential target in neurodegenerative disease e.g. Alzheimer and stroke [11, 20, 21]. One of the most frequently used p53-inhibitors is pifithrin-α (PFTα), a small molecule which was first described by Komarov and coworkers [22]. PFTα was shown to be beneficial in rodent models of PD [23, 24], epileptic seizures [25] and stroke [15, 20]. Although PFTα is claimed to be a specific inhibitor of p53, reports on PFTα interaction with the aryl hydrocarbon receptor [26], inhibition of nuclear translocation of p53 [27] and suppression of heat shock and glucocorticoid receptor signaling [28] question the specificity of PFTα.
Therefore, the aim of this study was to elucidate the effects of PFTα on cell viability and mitochondrial function in a model of glutamate-induced oxidative cell death in immortalized hippocampal HT-22 cells compared to a selective downregulation of p53 by using p53 siRNA. In HT-22 cells lacking NMDA-receptors, glutamate induces a depletion of glutathione and the subsequent accumulation of ROS which therefore results in lethal oxidative stress and mitochondrial fragmentation [4]. We report that PFTα provides neuroprotection in hippocampal HT-22 neurons by protecting mitochondria against glutamate-induced oxytosis independent of p53 inhibition. In contrast, p53 depletion only partly abolishes cell death and does not affect mitochondrial damage in intrinsic cell death triggered by oxidative stress.
Results
PFTα mediates neuroprotective effects against oxidative stress
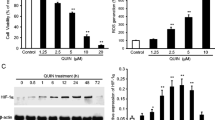
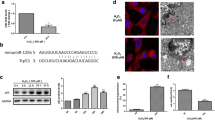
Millimolar concentrations of glutamate induce detrimental amounts of ROS in hippocampal HT-22 cells causing cell death 12–15 h after the glutamate challenge [5]. The damaged cells shrink, round up and detach from the bottom of the dish. Treatment with PFTα prevented these morphological changes completely (Fig. 1a). The morphological changes after glutamate exposure were also reflected in a loss of cell viability detected by the MTT assay. Co-treatment with PFTα rescued HT-22 cells from glutamate induced cell death in a concentration dependent manner (Fig. 1b). This neuroprotective effect of PFTα was also confirmed by Annexin V/PI staining and subsequent FACS analysis. In this assay, early apoptotic cells appear Annexin V-positive while late apoptotic and necrotic cells are Annexin V- and PI-positive [29]. Healthy cells remain unstained. Total cell death was determined by adding the values for both populations of dying cells showing positive staining for Annexin V and Annexin V/PI. Glutamate significantly increased the number of dead cells compared to control levels. The p53 inhibitor PFTα was able to abolish the increase in Annexin- and PI-positive cells after glutamate challenge almost to control level (Fig. 1c). In contrast, Pifithrin-µ (PFT-µ), another p53 inhibitor, which inhibits p53 binding to mitochondria [42], failed to protect HT-22 cells from glutamate induced toxicity. It even showed a dose-dependent toxicity (Fig. 1d). A post-treatment with PFTα up to 4 h after the glutamate challenge also significantly prevented the decrease in cell viability, but was not as protective as the simultaneous administration of glutamate and PFTα (Fig. 1e, f).
PFTα prevents glutamate-induced cell death. a PFTα (10 µM) preserves HT-22 cells from glutamate-induced (5 mM, 15 h) morphological changes compared to non-treated controls (×10). b MTT assay shows dose-dependent protection of PFTα against glutamate (7 mM, 16 h) toxicity. (###p < 0.001 compared to untreated control; *p < 0.05 and ***p < 0.001 compared to glutamate-treated control). c Neuroprotective effect of PFTα (10 µM) was determined by AnnexinV/PI staining (5 mM glutamate, 16 h). Data shown as mean + SD. (###p < 0.001 compared to untreated control; ***p < 0.001 compared to glutamate-treated control). d PFT-µ does not protect against glutamate toxicity (3 mM, 15 h). (###p < 0.001 compared to untreated control). e Impedance was measured in post-treatment conditions when PFTα (10 µM) was added 2, 4 and 6 h after the onset of glutamate (4 mM). PFTα attenuated cell death compared to glutamate treated controls when applied up to 4 h after the onset of glutamate (n = 7). f Bar graph evaluation at the 14 h time point from the xCELLigence recordings of Fig. 1e (right black arrow). (###p < 0.001 compared to untreated control; **p < 0.01 and ***p < 0.001 compared to glutamate-treated control)
PFTα preserves mitochondrial integrity
Mitochondrial damage is a key feature of cell death induced by oxidative stress in the present model system [4, 5, 30]. Hence, we examined mitochondrial morphology and functional hallmarks such as mitochondrial membrane potential (MMP) and ATP production. To this end, HT-22 cells were treated with glutamate and stained with TMRE, a lipophilic cationic dye indicating intact MMP by red fluorescence. Loss of MMP due to glutamate toxicity was determined by the loss of red fluorescence detected by FACS analysis. Carbonyl cyanide 3-chlorophenylhydrazone (CCCP), which acts as an uncoupler of the oxidative phosphorylation, was applied as a positive control for the loss of MMP. Glutamate did not affect the MMP to the same extend as CCCP, but induced a significant reduction of MMP compared to untreated control cells. PFTα preserved such MMP loss induced by glutamate almost at control levels. PFTα treatment itself did not influence the membrane potential at all (Fig. 2a). Next, we examined the mitochondrial morphology. As established in our earlier work, glutamate toxicity in HT-22 cells was accompanied by mitochondrial fragmentation [4, 30]. These fragmented mitochondria appear in a dotted pattern and are located in close vicinity to the nucleus of the damaged cells. Treatment with PFTα significantly prevented this glutamate-induced mitochondrial fragmentation. Mitochondria of PFTα–treated cells remained elongated and appeared as a tubular network all over the cell (Fig. 2b). Overall, these findings pointed to a protective mechanism of action of PFTα at the level of mitochondria.
PFTα preserves MMP and morphology. a MMP measured by TMRE fluorescence is fully restored by PFTα (10 µM) after 3 mM glutamate, 14 h. Data shown as mean + SD. (###p < 0.001 compared to untreated control; ***p < 0.001 compared to glutamate-treated control). b Fluorescence pictures show conservation of mitochondrial morphology after PFTα (10 µM) in combination with glutamate (5 mM, 14 h). Cells were stained with Mitotracker green and DAPI. Scale bar 20 µm. c Quantification of mitochondrial morphology; Category I (Cat I): elongated, Category II (Cat II): intermediate, Category III (Cat III): fragmented. Values are given from five independent experiments with at least 500 cells per condition counted blind. (###p < 0.001 compared to CatI of untreated control; ***p < 0.001 compared to Cat I of glutamate-treated control). Statistics were obtained using ANOVA, Scheffé Test
PFTα blocks lipid peroxidation
Lipid peroxidation is a well-established trigger mechanism of mitochondrial demise and neuronal cell death in the present model of oxytosis induced by glutamate in HT-22 cells [5, 31, 32]. Accordingly, we detected lipid peroxidation using fluorescent Bodipy staining and subsequent FACS analysis. Bodipy fluorescence is shifted from red to green upon oxidation. At both time points, at 7 and 15 h after the glutamate challenge, PFTα attenuated the formation of lipid peroxides in a concentration-dependent manner (Fig. 3a, b) which correlated well with the previous results from the MTT assays where the protective effect also showed a concentration dependency (Fig. 1b).
PFTα reduces lipid peroxidation. a PFTα lowers lipid peroxidaton after glutamate challenge (7 mM, 15 h) in a concentration dependent manner detected by Bodipy staining and FACS analysis. PFTα was added 1 h before glutamate treatment. b Bodipy staining after 7 h glutamate (7 mM) and PFTα pretreatment. Data shown as mean + SD. (###p < 0.001 compared to untreated control; ***p < 0.001 and *p < 0.05 compared to glutamate-treated control)
Selective p53 silencing attenuates glutamate-induced toxicity
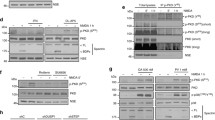
PFTα is a widely used inhibitor of p53 [22]. To link our previous results to this proposed inhibition we investigated the effects of a siRNA-mediated downregulation of p53 protein levels in HT-22 cells on cell viability and mitochondrial integrity. The knockdown of p53 was achieved by siRNA transfection and confirmed at mRNA and protein levels using RT-PCR and Western Blot analysis, respectively (Fig. 4a). Gene silencing of p53 attenuated glutamate-induced cytotoxicity as detected by the MTT assay while transfection of controls with the scrambled siRNA did not alter cell viability. PFTα co-treatment was used as a positive control for cell survival and appeared more potent than p53 downregulation (Fig. 4b). To rule out off target effects we confirmed these observations by using another p53 siRNA as shown in the supplements (online resource 1). As the MTT assay is just an endpoint measurement, we also detected cell viability over time using the real time impedance measurement system (xCELLigence). Here we could show a delay of cell death by selective p53 silencing for about 2 h compared to cells transfected with a non-specific scrambled siRNA (Fig. 4c) which could explain a lesser effect than in comparison to PFTα in the MTT. PFTα administration to p53 silenced cells was still protective against glutamate induced cytotoxicity and even more than p53 silencing alone (Fig. 4d, e).
Selective p53 knockdown attenuates glutamate toxicity. a Knockdown of p53 by siRNA (20 nM) was verified by RT-PCR and Western Blot analysis. Quantification of 7 independent Western Blots shows a significant reduction of p53 protein level. (***p < 0.001 compared to scrambled siRNA). b MTT assay shows increased cell viability of cells transfected with p53 siRNA (20 nM) compared to cells transfected with scrambled control siRNA (20 nM) after glutamate challenge (3 mM, 14 h). (###p < 0.001 compared to untreated scrambled siRNA; ***p < 0.001 compared to glutamate-treated scrambled siRNA). c xCELLigence real-time measurement: Cells transfected with p53 siRNA (20 nM) show a short transient protection against treatment with 3 mM glutamate (glut) compared to cells transfected with scrambled siRNA. d xCELLigence real-time measurement reveals protection of knocking down p53 and PFTα treatment at the same time against glutamate (5 mM) toxicity. e Bar graph evaluation at the 15 h time point from the xCELLigence recordings of Fig. 4d (right black arrow)
p53 gene silencing fails to preserve mitochondrial integrity after the glutamate challenge
As p53 silencing had similar but only transient protective effects on cell viability compared to PFTα treatment, we further investigated the impact of p53 knockdown on mitochondrial integrity. The FACS analysis of cells stained with TMRE revealed that p53 siRNA-mediated knockout failed to prevent mitochondrial depolarization after the glutamate challenge for 14 h (Fig. 5a). Transfection of p53 siRNA in control cells did not change MMP at all. After 7 h of glutamate treatment there were no changes in the MMP detectable in cells transfected with scrambled siRNA as well as in p53 silenced cells (Fig. 5a). Further, after the glutamate challenge we observed similar increases in mitochondrial fragmentation (category II and III) and decreasing percentages of tubular mitochondria (category I) in cells treated either with scrambled siRNA or p53 targeting siRNA, suggesting that the depletion of p53 did not prevent mitochondrial fragmentation. Notably, siRNA transfection itself did not alter the shape of the mitochondria in control cells which predominantly contained elongated and tubular mitochondria distributed in a tubular network all over the cell (Fig. 5b).
p53 siRNA does not keep mitochondrial integrity. a MMP was determined after 7 and 14 h glutamate (3 mM). CCCP was used as positive control for breakdown of MMP. b Fluorescence photomicrographs show categories of mitochondrial morphology: Category I (Cat I): elongated, Category II (Cat II): intermediate, Category III (Cat III): fragmented; scale bar 20 µm. Cells were stained with Mitotracker Red before glutamate challenge (3 mM, 14 h) and quantified by counting at least 500 cells per condition blind to treatment
p53 knockdown blocks p53 transcriptional activity
It is well established that p53 can induce cell death through induction of different proapoptotic target genes [33]. It has recently been shown that the inhibition of PUMA and NOXA upregulation by PFT-µ was associated with reduced cerebral damage in a model of hypoxic-ischemic brain damage. In contrast to that, the beneficial effect of PFTα in this model was much smaller although it fully blocked p53 transcriptional activity [19]. To address the question, whether downregulation of p53 was also able to diminish p53 transcriptional activity in HT-22 cells despite the small effect on cell viability, we created a p53-Luc reporter plasmid. This plasmid was transfected into p53 silenced HT-22 cells and p53 transcriptional activity was assessed in the presence or absence of glutamate to measure luciferase activity. As expected, siRNA-mediated p53 gene silencing reduced p53 transcriptional activity compared to the controls. Unfortunately, this assay is not suitable to investigate the described inhibitory effect of PFTα on p53 transcriptional activity because a direct inhibition of firefly luciferase has been reported for PFTα concentrations higher than 1 µM [34], and this unspecific inhibition of luciferase activities was also confirmed in our current experimental setup (data not shown). To further compare the effects of pharmacological p53 inhibition and selective p53 gene silencing on transcriptional p53 activity, we determined the mRNA expression levels of p53 target genes known to be associated with mitochondrial function and regulation of apoptosis. Here, we confirmed the functionality of the p53 siRNA on p53 mRNA expression, while PFTα did not affect p53 mRNA levels. The amount of the proapoptotic proteins AIF, DRP1, PUMA and Bid remained unchanged after treatment with both, PFTα or p53 siRNA, in the presence or absence of glutamate. Notably, the induction of oxytosis by glutamate itself was not accompanied by changes in the mRNA level of any of these proteins. The mRNA levels of the negative p53 regulator MDM2 were decreased in cells with p53 silencing but not in the presence of PFTα. Further, we found a downregulation of the cell cycle regulator p21 after p53 knockout both with and without induction of oxidative stress, whereas this was not observed with PFTα treatment (Fig. 6b, c).
p53 siRNA blocks p53 transcriptional activity. a p53 silencing diminishes p53 transcriptional activity. Reporter assay was performed after glutamate challenge (3 mM, 14.5 h). Data shown as mean + SD. (***p < 0.001 compared to untreated vehicle; ###p < 0.001 compared to glutamate-treated vehicle.). b RT-PCR analysis of p53 target genes after PFTα-treatment and glutamate challenge (7 mM, 16 h). c RT-PCR analysis of p53 target genes after p53 knockout and glutamate challenge (7 mM, 15 h)
Discussion
The present study shows that the inhibition of p53 protects neuronal HT-22 cells against glutamate-induced oxytosis. Regarding cell viability, PFTα was more potent than p53 silencing via RNA interference. The observed protection of HT-22 cells by both, PFTα-mediated pharmacological p53 inhibition and p53 gene silencing, is in line with findings reported in other model systems of neuronal cell death in the literature. Yonekura and coworkers observed reduced neuronal death in p53−/− mice after global cerebral ischemia compared to p53+/+mice [35]. Further, in a model of permanent middle cerebral artery occlusion (pMCAO), p53 knockout reduced infarct size [36]. PFTα also reduces contusion volume in mice after traumatic brain injury [16]. Considering these findings one would not expect such a notable difference between p53 gene silencing and pharmacological p53 inhibition with PFTα regarding cell viability as described in the present study. It should be considered that we do not achieve a sustained full knock out of p53 levels and transcriptional activity using RNA interference while a concentration of 10 µM PFTα is rather high and probably able to fully block p53 action leading to a more pronounced effect, particularly at the level of mitochondria which would explain our observations.
Differential effects of pharmacological p53 inhibition and genetic knockout were also described in previous studies. For example, in a model of Pol I-specific transcription initiation factor IA (TIF-IA) depleted dopaminergic neurons the inhibition of p53 by PFTα decreased the amount of apoptotic cells [37] while in TIF-IA depleted medium spiny neurons p53 knockout led to an increase of apoptotic cells [38]. Mendjargal and coworkers also demonstrated in a murine macrophage-like cell line that a siRNA-mediated down regulation of p53 did not affect lipopolysaccharide (LPS)-induced iNOS expression whereas PFTα strongly reduced iNOS levels after LPS treatment also in p53-silenced cells which points towards a p53-independent action of PFTα in this model system as well [39].
In contrast to inhibition of p53 by PFTα the gene silencing of p53 was not able to preserve mitochondrial integrity after the glutamate challenge. Thus, the present data suggest an action of PFTα upstream of mitochondria preventing glutamate-induced oxytosis indicated by a pronounced protection of MMP and mitochondrial morphology (Fig. 2). This is in line with earlier findings in cultured primary neurons [20, 40] and in synaptosomes [40] where PFTα also preserved mitochondrial function after insults induced by DNA damage, glutamate or oxidative stress.
Our diverse findings regarding effects of PFTα and p53-siRNA on mitochondria are important to explain the differences in the respective effects on cell viability. Our findings indicate pleiotropic effects of PFTα beyond the inhibition of p53 that mediate protection of neurons against oxidative stress. This conclusion is supported by previous findings on interactions of PFTα with the aryl hydrocarbon receptor [26] or HSP70 [28]. Further, PFTα may also induce mitochondrial protection and neuroprotection in ischemic brain tissue via activation of Akt and enhanced NF-κB signaling [15, 41]. The beneficial effects of PFTα on mitochondria and the sustained neuroprotective effect compared to mere p53 silencing detected in the present study underline once more the pivotal role of mitochondrial integrity for neuroprotection.
The full mitochondrial protection is also accompanied by the reduction of lipid peroxide formation in the presence of PFTα (Fig. 3). This stands in contrast to the observations of Nijboer et al. who did not see a reduction in lipid peroxidation in a model of hypoxic-ischemic brain injury by PFTα, but by PFT-µ [19] which inhibits mitochondrial association of p53 [42], thus mediating its protective effect. However, PFT-µ also failed to protect HT-22 cells against glutamate-induced oxytosis (Fig. 1d) supporting our hypothesis of a p53-independent mitochondrial mechanism of action of PFTα in the present model system of oxytosis.
In order to gain deeper insights into the mechanisms of protection at the level of mitochondria we screened for several target genes of p53 that were previously associated with mitochondrial pathways of cell death. Although there are numerous target genes of p53 involved in cell death it is still controversially discussed which of these are the most important for p53-mediated cell death [33]. So far, it seems to depend on the kind of stress and cell type which target gene is the most relevant for p53-dependent induction of apoptosis [43].
MDM2 is one of the major regulators of p53 activity and also one of its transcriptional targets acting in a negative feedback loop [33] thereby controlling p53 levels during normal growth [44]. This described negative regulation is consistent with our observed decrease in MDM2 expression levels after siRNA-mediated p53 gene silencing (Fig. 6c).
While we could not observe any differences in expression levels of AIF, DRP1, Bid or PUMA after both, PFTα treatment or p53 downregulation, the most striking effect was the decrease of p21 levels after p53 silencing in the presence as well as in the absence of glutamate. p21 is the major p53-dependent mediator of cell cycle arrest [10, 33]. Yu et al. showed that p21−/− cancer cells are more susceptible to p53-induced apoptosis than wild type cells [45, 46] leading to the conclusion that in the present model system less p21 attributed to p53 gene silencing may render the HT-22 cells more sensitive to the imposed oxidative stress. In contrast, Leker and colleagues claimed that a reduction of p21 by PFTα decreased the amount of apoptotic cells in ischemic brain tissue due to the inhibition of p53 transcriptional activity [47]. In the present study the reduced transcriptional activity of p53 in p53 silenced cells (Fig. 6a) led to a decline of p21 expression levels which was not found after PFTα treatment (Fig. 6b, c). Together with the differential outcome on cell viability after p53 gene silencing versus the effects of PFTα, in the present setting of experiments lower levels of p21 correlated with enhanced susceptibility of the cells for oxidative stress.
PUMA is one of the most prominent pro-apoptotic p53 target genes which act at the level of mitochondria in p53-dependent apoptosis [48]. Recent studies showed an upregulation of PUMA in damaged neurons after transient global cerebral ischemia [49] and after status epilepticus triggered by intra-amygdala kainic acid in mice [25], these effects were abolished by PFTα. As there is no change in PUMA expression in our model we can conclude that glutamate-induced oxytosis in HT-22 cells is not reliant on p53-dependent transcriptional activation of PUMA, which is also supported by the limited effect of p53 downregulation on cell survival compared to the pronounced and sustained protection achieved by PFTα. According to previous results in cancer cells, conserved PUMA levels and depletion of p21 [50] are required for mitochondrial damage and subsequent cell death while disruption of both PUMA and p21 prevented p53-induced apoptosis [45]. Since p53 gene silencing resulted in a similar expression pattern of reduced p21 levels and stable PUMA levels, this may also explain why p53 silencing failed to protect the HT-22 cells more efficiently.
In the present study, we showed a strong neuroprotective effect of the p53 inhibitor PFTα in HT-22 cells contrary to p53 gene silencing which only had partial effects on neuronal viability in a model of oxidative stress and did not affect mitochondrial integrity at all. The current data suggest a protective mechanism of PFTα upstream or at the level of mitochondria which must be either independent of p53 inhibition or accompanied by additional effects which could be the subject of future studies. All in all our results again confirm the neuroprotective potential of PFTα in neurodegenerative disease associated with oxidative stress and the importance of a well controlled p53 expression.
Materials and methods
Cells
HT-22 cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Invitrogen, Karlsruhe, Germany) with the addition of 10 % heat-inactivated fetal calf serum, 100 U/ml penicillin, 100 mg/ml streptomycin and 2 mM glutamine (all Sigma-Aldrich, Munich, Germany).
For inducing cell death, 3–7 mM glutamate (Sigma-Aldrich, Munich, Germany) was added to the medium for the indicated amount of time.
SiRNA transfections were performed by using Lipofectamine RNAiMax (Invitrogen, Karlsruhe, Germany) following the manufacturers protocol. After complex formation, an adequate number of cells were added in antibiotic-free medium to the transfection mix according to the following experimental procedure. Cells were treated after growing for 48 h. The following siRNA sequences were used: CCACUUGAUGGAGAGUAUU (p53 siRNA), UAAUGUAUUGGAACGCAUA (scrambled siRNA).
Pifithrin-α (Sigma-Aldrich, Munich, Germany) was dissolved in DMSO and used in a concentration of 10 µM as pretreatment (1 h) and cotreatment with glutamate.
Pifithrin-µ (Sigma-Aldrich, Munich, Germany) was dissolved in DMSO.
Cell viability
Cell viability was quantified by the reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) for 1 h at 37 °C. The dye was dissolved in an appropriate amount of DMSO and absorbance measured at 570 nm versus 630 nm with FluoStar (BMG Labtech, Offenburg, Germany).
For real time measurement of cell viability, the xCELLigence system (Roche, Penzberg, Germany) was used as described earlier [51].
Additionally, cell viability was detected by an Annexin V/PI staining using an Annexin-V-FITC Detection Kit (Promokine, Heidelberg, Germany) followed by fluorescence-activated cell sorting (FACS) analysis. Data were collected from 10,000 cells from at least four wells per condition.
Mitochondrial membrane potential
MMP was measured by FACS analysis using a MitoPT™ TMRE Kit (Immunochemistry Technologies, Hamburg, Germany). Cells were treated with glutamate for 7–15 h and harvested by trypsin. After staining for 20 min with tetramethylrhodamin ethyl ester (TMRE) at 37 °C, cells were washed with PBS and resuspended in assay buffer. Data were collected from 10,000 cells from at least four wells per condition.
Mitochondrial morphology
For analyzing mitochondrial morphology, cells were stained with Mitotracker red/green and DAPI. After glutamate treatment, cells were fixed with 4 % paraformaldehyde. At least 500 cells per condition were counted blind to treatment. Images were acquired using a fluorescence microscope (Leica, Wetzlar, Germany).
Lipid peroxidation
The formation of lipid peroxides was determined by staining the cells with 2 µM BODIPY 581/591 C11 (Invitrogen) for 1 h at 37 °C in culture medium after glutamate treatment at indicated time points. After collecting the cells, flow cytometric analysis was done in PBS with an excitation at 488 nm and an emission at 530 and 585 nm. Data were collected from 10,000 cells from at least four wells per condition.
Protein analysis and Western Blotting
For protein analysis, cells were treated as indicated. After washing with PBS, cells were lysed with buffer containing 0.25 M Mannitol, 0.05 M Tris, 1 M EDTA, 1 M EGTA, 1 mM DTT, 1 % Triton-X (all Sigma-Aldrich), supplemented with Complete Mini Protease Inhibitor Cocktail and PhosSTOP (both Roche Diagnostics, Penzberg, Germany). To eliminate membrane fragments and other insoluble components, extracts were centrifuged at 13000xg for 15 min at 4 °C. The amount of protein was measured by Pierce® BCA Protein Assay Kit (Perbio Science, Bonn, Germany).
For Western Blot analysis, samples were run on a 12.5 % SDS-Gel and blotted onto a PVDF-membrane at 15 V for 90 min. Blots were incubated with primary antibody at 4 °C overnight. The following primary antibodies were used: Anti-p53 (1C12) (Cell Signaling, Danvers, MA, USA) and Anti-Actin C4 (MB Biomedicals, Illkirch Cedex, France). After incubation with proper secondary HRP-labeled antibody (Vector Laboratories, Burlingame, CA, USA) Western Blot signals were detected by chemiluminescence with Chemidoc software (Bio-Rad, Munich, Germany).
RT-PCR
Total RNA amount was extracted after indicated treatment by InviTrap® Spin Universal RNA Kit (Stratec molecular, Berlin, Germany). RT-PCR was performed with SuperScript III One-Step RT-PCR Kit with Platinum Taq (Invitrogen). Following primers were used p53: forward 5′-GACCGCCGTACAGAAGAAGA-3′ and reverse 5′-GCCCCACTTTCTTGACC-3′; GAPDH: forward 5′-AGGCCGGTGCTGAGTAT-3′ and reverse 5′-TGCCTGCTTCACCACC TTCT-3′; AIF: forward 5′-GCGTAATACGACTCACTATAGGGAGATCCAGGCAACTTGT TCCAGC-3′ and reverse 5′-CGTAATACGACTCACTATAGGGAGACCTCTGCTCCAGCC CTATCG-3′; p21 forward 5′-GCAGATCCACAGCGATATCC-3′ and reverse 5′-CAACTGC TCACTGTCCACGG-3′; DRP1 forward 5′-ACAGGAGAAGAAAATGGAGTTTGAAG CAG-3′ and reverse 5′-AACAAATCCTAGCACCACGCAT-3′; Bid forward 5′-GGGA ACTGCCTGTGCAAGCTTAC-3′ and reverse 5′-CAGTGAGGFCCTTGTCTCTGAA-3′; MDM2 forward 5′-CCAGCTTCGGAACAAGAGAC-3′ and reverse 5′-ACACAAT GTGCTGCTGCTTC-3′; PUMA forward 5′-CAGACTGTGAATCCTGTGCT-3′ and reverse 5′-ACAGTATCTTACAGGCTGGGG-3′. For visualization of the DNA products, UV illumination on a 1.5 % agarose gel was used.
Luciferase-reporter-assay
To investigate the effect of p53 siRNA on p53 transcriptional activity we have generated a luciferase reporter gene construct harboring six consensus p53 DNA binding elements [52]. The oligonucleotides containing p53 DNA binding elements used in this study were sense 5′-CTAGCAGACATGCCTAGACATGCCTAGACATGCCTAGACATGCCTAGACATGCCT AGACATGCCTCGA-3′ and anti-sense 5′-GATCTCGAGGCATGTCTAGGCATGTCTAGGCATG TCTAGGCATGTCTAGGCATGTCTAGG CATGTCTG-3′ and were flanked by sites for Nhe1 and Bgl2 restriction enzymes. One microgram of each oligonucleotide was mixed together and heated at 95 °C for 5 min in a water bath and allowed to cool slowly to room temperature. These annealed oligonucleotides were ligated into Nhe1 and Bgl2 restriction sites of the pTAL-Luciferase (Luc) reporter plasmid yielding p53-Luc reporter vector. pTAL-Luc vector contains a TATA-like promoter region from the Herpes simplex virus thymidine kinase (HSV-TK) promoter to drive the luciferase gene expression upon p53 binding to its enhancer element.
For detection of p53-transcriptional activity, cells were transfected with p53 siRNA as previously described. After 1 day, Renilla-plasmid and p53-Luc-plasmid were cotransfected using Attracten® (Qiagen, Hilden, Germany). Twenty-four hours later cells were treated as indicated and luciferase assay was performed with Dual-Luciferase® Assay Kit (Promega, Madison, USA) following manufacturers protocol. Luminescence was measured with FluoStar (BMG Labtech, Offenburg, Germany).
Statistical analysis
All data are given as mean + standard deviation (SD). Statistical comparison between treatment groups was performed by analysis of variance (ANOVA) followed by Scheffé’s post hoc test. Calculations were executed with Winstat standard statistical software (R. Fitch Software, Bad Krozingen, Germany).
Abbreviations
- AIF:
-
Apoptosis inducing factor
- AD:
-
Alzheimer’s disease
- ANOVA:
-
Analysis of variance
- Bid:
-
BH3 interacting-domain death agonist
- CCCP:
-
Carbonyl cyanide 3-chlorophenylhydrazone
- DRP1:
-
Dynamin-related protein 1
- FACS:
-
Fluorescence-activated cell sorting
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- LPS:
-
Lipopolysaccharide
- MDM2:
-
Mouse double minute 2 homolog
- MMP:
-
Mitochondrial membrane potential
- MTT:
-
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PCD:
-
Programmed cell death
- PD:
-
Parkinson’s disease
- PFTα:
-
Pifithrin-α
- pMCAO:
-
Permanent middle cerebral artery occlusion
- PUMA:
-
p53 upregulated modulator of apoptosis
- ROS:
-
Reactive oxygen species
- TIF-IA:
-
Pol I-specific transcription initiation factor IA
- TMRE:
-
Tetramethylrhodamin ethyl ester
References
Culmsee C, Landshamer S (2006) Molecular insights into mechanisms of the cell death program: role in the progression of neurodegenerative disorders. Curr Alzheimer Res 3(4):269–283
Mattson MP (1998) Modification of ion homeostasis by lipid peroxidation: roles in neuronal degeneration and adaptive plasticity. Trends Neurosci 21(2):53–57
Lin MT, Beal MF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443(7113):787–795
Grohm J, Plesnila N, Culmsee C (2010) Bid mediates fission, membrane permeabilization and peri-nuclear accumulation of mitochondria as a prerequisite for oxidative neuronal cell death. Brain Behav Immun 24(5):831–838
Tobaben S, Grohm J, Seiler A, Conrad M, Plesnila N, Culmsee C (2011) Bid-mediated mitochondrial damage is a key mechanism in glutamate-induced oxidative stress and AIF-dependent cell death in immortalized HT-22 hippocampal neurons. Cell Death Differ 18(2):282–292
Galluzzi L, Blomgren K, Kroemer G (2009) Mitochondrial membrane permeabilization in neuronal injury. Nat Rev Neurosci 10(7):481–494
Zhang J, Cao Q, Li S, Lu X, Zhao Y, Guan J, Chen J, Wu Q, Chen G (2013) 3-hydroxybutyrate methyl ester as a potential drug against Alzheimer’s disease via mitochondria protection mechanism. Biomaterials 34(30):7552–7562
Marques-Aleixo I, Oliveira PJ, Moreira PI, Magalhães J, Ascensão A (2012) Physical exercise as a possible strategy for brain protection: evidence from mitochondrial-mediated mechanisms. Prog Neurobiol 99(2):149–162
Checler F, da Costa CA (2014) p53 in neurodegenerative diseases and brain cancers. Pharmacol Ther 142(1):99–113
Tomasevic G, Shamloo M, Israeli D, Wieloch T (1999) Activation of p53 and its target genes p21WAF1/Cip1 and PAG608/Wig-1 in ischemic preconditioning. Mol Brain Res 70(2):304–313
Culmsee C, Mattson MP (2005) p53 in neuronal apoptosis. Biochem. Biophys. Res. Commun. 331(3):761–777
Vousden KH, Prives C (2009) Blinded by the light: the growing complexity of p53. Cell 137(3):413–431
Muller Patricia A J, Vousden KH (2013) p53 mutations in cancer. Nat Cell Biol 15(1):2–8
Xiang H, Hochman DW, Saya H, Fujiwara T, Schwartzkroin PA, Morrison RS (1996) Evidence for p53-mediated modulation of neuronal viability. J Neurosci 16(21):6753–6765
Culmsee C, Siewe J, Junker V, Retiounskaia M, Schwarz S, Camandola S, El-Metainy S, Behnke H, Mattson MP, Krieglstein J (2003) Reciprocal inhibition of p53 and nuclear factor-kappaB transcriptional activities determines cell survival or death in neurons. J Neurosci 23(24):8586–8595
Plesnila N, von Baumgarten L, Retiounskaia M, Engel D, Ardeshiri A, Zimmermann R, Hoffmann F, Landshamer S, Wagner E, Culmsee C (2007) Delayed neuronal death after brain trauma involves p53-dependent inhibition of NF-kappaB transcriptional activity. Cell Death Differ 14(8):1529–1541
Green DR, Kroemer G (2009) Cytoplasmic functions of the tumour suppressor p53. Nature 458(7242):1127–1130
Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM (2012) p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149(7):1536–1548
Nijboer CH, Heijnen CJ, van der Kooij MA, Zijlstra J, van Velthoven CTJ, Culmsee C, van Bel F, Hagberg H, Kavelaars A (2011) Targeting the p53 pathway to protect the neonatal ischemic brain. Ann Neurol 70(2):255–264
Culmsee C, Zhu X, Yu QS, Chan SL, Camandola S, Guo Z, Greig NH, Mattson MP (2001) A synthetic inhibitor of p53 protects neurons against death induced by ischemic and excitotoxic insults, and amyloid beta-peptide. J Neurochem 77(1):220–228
Zhu G, Wang X, Wu S, Li Q (2012) Involvement of activation of PI3K/Akt pathway in the protective effects of puerarin against MPP+-induced human neuroblastoma SH-SY5Y cell death. Neurochem Int 60(4):400–408
Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV (1999) A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 285(5434):1733–1737
Duan W, Zhu X, Ladenheim B, Yu Q, Guo Z, Oyler J, Cutler RG, Cadet JL, Greig NH, Mattson MP (2002) p53 inhibitors preserve dopamine neurons and motor function in experimental parkinsonism. Ann Neurol 52(5):597–606
Chou J, Greig NH, Reiner D, Hoffer BJ, Wang Y (2011) Enhanced survival of dopaminergic neuronal transplants in hemiparkinsonian rats by the p53 inactivator PFT-α. Cell Transpl 20(9):1351–1359
Engel T, Murphy BM, Hatazaki S, Jimenez-Mateos EM, Concannon CG, Woods I, Prehn JHM, Henshall DC (2010) Reduced hippocampal damage and epileptic seizures after status epilepticus in mice lacking proapoptotic Puma. FASEB J 24(3):853–861
Hoagland MS, Hoagland EM, Swanson HI (2005) The p53 inhibitor pifithrin-alpha is a potent agonist of the aryl hydrocarbon receptor. J Pharmacol Exp Ther 314(2):603–610
Murphy PJM, Galigniana MD, Morishima Y, Harrell JM, Kwok RPS, Ljungman M, Pratt WB (2004) Pifithrin-alpha inhibits p53 signaling after interaction of the tumor suppressor protein with hsp90 and its nuclear translocation. J Biol Chem 279(29):30195–30201
Komarova EA, Neznanov N, Komarov PG, Chernov MV, Wang K, Gudkov AV (2003) p53 inhibitor pifithrin alpha can suppress heat shock and glucocorticoid signaling pathways. J Biol Chem 278(18):15465–15468
Schutte B, Nuydens R, Geerts H, Ramaekers F (1998) Annexin V binding assay as a tool to measure apoptosis in differentiated neuronal cells. J Neurosci Methods 86(1):63–69
Grohm J, Kim S, Mamrak U, Tobaben S, Cassidy-Stone A, Nunnari J, Plesnila N, Culmsee C (2012) Inhibition of Drp1 provides neuroprotection in vitro and in vivo. Cell Death Differ 19(9):1446–1458
Tan S, Sagara Y, Liu Y, Maher P, Schubert D (1998) The regulation of reactive oxygen species production during programmed cell death. J Cell Biol 141(6):1423–1432
Li Y, Maher P, Schubert D (1997) A role for 12-lipoxygenase in nerve cell death caused by glutathione depletion. Neuron 19(2):453–463
Vousden KH (2000) p53: death star. Cell 103(5):691–694
Rocha S, Campbell KJ, Roche KC, Perkins ND (2003) The p53-inhibitor pifithrin-alpha inhibits firefly luciferase activity in vivo and in vitro. BMC Mol Biol 4:9
Yonekura I, Takai K, Asai A, Kawahara N, Kirino T (2006) p53 potentiates hippocampal neuronal death caused by global ischemia. J Cereb Blood Flow Metab 26(10):1332–1340
Crumrine RC, Thomas AL, Morgan PF (1994) Attenuation of p53 expression protects against focal ischemic damage in transgenic mice. J Cereb Blood Flow Metab 14(6):887–891
Rieker C, Engblom D, Kreiner G, Domanskyi A, Schober A, Stotz S, Neumann M, Yuan X, Grummt I, Schütz G, Parlato R (2011) Nucleolar disruption in dopaminergic neurons leads to oxidative damage and parkinsonism through repression of mammalian target of rapamycin signaling. J Neurosci 31(2):453–460
Kreiner G, Bierhoff H, Armentano M, Rodriguez-Parkitna J, Sowodniok K, Naranjo JR, Bonfanti L, Liss B, Schütz G, Grummt I, Parlato R (2013) A neuroprotective phase precedes striatal degeneration upon nucleolar stress. Cell Death Differ 20(11):1455–1464
Mendjargal A, Odkhuu E, Koide N, Nagata H, Kurokawa T, Nonami T, Yokochi T (2013) Pifithrin-α, a pharmacological inhibitor of p53, downregulates lipopolysaccharide-induced nitric oxide production via impairment of the MyD88-independent pathway. Int Immunopharmacol 15(4):671–678
Gilman CP, Chan SL, Guo Z, Zhu X, Greig N, Mattson MP (2003) p53 is present in synapses where it mediates mitochondrial dysfunction and synaptic degeneration in response to DNA damage, and oxidative and excitotoxic insults. Neuromol Med 3(3):159–172
Duan W, Li Q, Xia M, Tashiro S, Onodera S, Ikejima T (2011) Silibinin activated p53 and induced autophagic death in human fibrosarcoma HT1080 cells via reactive oxygen species-p38 and c-Jun N-terminal kinase pathways. Biol Pharm Bull 34(1):47–53
Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, Bosykh DA, Burdelya LG, Macklis RM, Skaliter R, Komarova EA, Gudkov AV (2006) Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol 2(9):474–479
Beckerman R, Prives C (2010) Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2(8):a000935
Toledo F, Wahl GM (2006) Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 6(12):909–923
Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L (2003) PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci USA 100(4):1931–1936
Yu J, Zhang L (2005) The transcriptional targets of p53 in apoptosis control. Biochem Biophys Res Commun 331(3):851–858
Leker RR, Aharonowiz M, Greig NH, Ovadia H (2004) The role of p53-induced apoptosis in cerebral ischemia: effects of the p53 inhibitor pifithrin alpha. Exp Neurol 187(2):478–486
Nakano K, Vousden KH (2001) PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 7(3):683–694
Niizuma K, Endo H, Nito C, Myer DJ, Chan PH (2009) Potential role of PUMA in delayed death of hippocampal CA1 neurons after transient global cerebral ischemia. Stroke 40(2):618–625
Jansson M, Durant ST, Cho E, Sheahan S, Edelmann M, Kessler B, La T, Nicholas B (2008) Arginine methylation regulates the p53 response. Nat Cell Biol 10(12):1431–1439
Diemert S, Dolga AM, Tobaben S, Grohm J, Pfeifer S, Oexler E, Culmsee C (2012) Impedance measurement for real time detection of neuronal cell death. J Neurosci Methods 203(1):69–77
Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B (1997) 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell 1(1):3–11
Acknowledgments
We thank the excellent technical support by Mrs. Katharina Elsässer and Eileen Daube. Moreover, we thank Mrs. Emma Esser for careful editing of the manuscript and Roche Diagnostics GmbH for providing support with the xCELLigence system.
Conflict of interest
The authors declare that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Neitemeier, S., Ganjam, G.K., Diemert, S. et al. Pifithrin-α provides neuroprotective effects at the level of mitochondria independently of p53 inhibition. Apoptosis 19, 1665–1677 (2014). https://doi.org/10.1007/s10495-014-1048-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-014-1048-2