Abstract
Rhipicephalus microplus is a hematophagous ectoparasite that significantly affects parasitized cattle. As a one-host tick its entire life cycle consists of free-living and parasitic forms. Its extraordinary ability to survive during prolonged off-host periods has been related to the process of cytoplasmic degradation called autophagy. In order to deepen our understanding of this process during R. microplus non-parasitic stages, we determined the expression dynamics of a set of five autophagy-related genes (ATG genes) during embryonic development and over an increasing larval starvation period of 50 days. We found two apparent successive waves of ATG genes transcriptional activation, which paralleled key embryonic changes such as cellularization and organogenesis, as well as nutrient utilization. Moreover, during increasing larval starvation, ATG genes were up-regulated cyclically every 10–15 days. Taken together, our results suggest that autophagy is playing a major role in embryo development and energy metabolism during starvation in R. microplus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The tick Rhipicephalus microplus is an obligatory hematophagous ectoparasite that causes major losses to bovine herds. R. microplus is a one-host tick and its life cycle consists of the free-living and the parasitic forms. The free-living form is composed of engorged female ticks dropped off the host to the ground, laid eggs and free-living larvae, whereas the parasitic form is characterized by larval, nymphal and adult development on a single bovine host (Nuñez et al. 1985). Tick embryo development and survival of free-living larvae depend exclusively on maternal nutrients packaged into the oocytes during the pre-oviposition phase (Campos et al. 2006). While embryonic development under laboratory conditions lasts approximately 24 days (Davey et al. 1980; Senbill et al. 2018), the host-seeking period for eclosioned larvae can last from a few days to many months (Nuñez et al. 1985). This exceptional ability of ticks to survive for extended periods without food has been related to the process of intracellular protein degradation called autophagy. In order to advance our understanding of autophagy in R. microplus, during non-parasitic stages, in the present study we determined the expression dynamics of ATG genes reported so far for this tick, namely, RmATG3, RmATG4, RmATG6, RmATG8a and RmATG8b, whose products participate in the autophagosome formation and maturation (Flores et al. 2014, 2016), during embryonic development and over increasing larval starvation. We propound potential connections between ATG genes transcriptional activation, developmental changes and energy metabolism.
Materials and methods
Ticks and sample collection
Engorged adult female R. microplus ticks were collected from naturally infested cattle in Tapalpa, Jalisco, México. Ticks were washed in distilled water, placed into Petri dishes and incubated at 28 °C and 85% relative humidity (RH) throughout the egg-laying period. The length of the oviposition period averaged 14 days. Dead specimens were daily retired from dishes. Eggs (1 g per sample) were daily transferred to glass tubes with cotton plugs and incubated under the above-mentioned conditions. Eggs were collected and processed at 0, 5, 10, 15 and 20 days after oviposition (these time points covered the entire embryonic development period). Larvae were collected and processed at 0, 5, 10, 15, 20, 25, 30, 35, 40, 45 and 50 days after hatching. These time points are representative of an increasing larval starvation stress. It is worth mentioning that all the eggs and larvae evaluated in the study came from different females.
RNA isolation and cDNA synthesis
Total RNA was extracted from eggs and larvae immediately after collection using RNeasy Mini Kit (Qiagen, Germany) and following manufacturer’s recommendations. Briefly, 100 mg of eggs (~ 2000 eggs) or larvae (~ 1000 larvae) were independently mixed and crushed using a pellet pestle homogenizer. RNA integrity was determined by denaturing electrophoresis on 1% agarose gels. RNA was quantified by using a ND-1000 NanoDrop spectrophotometer (NanoDrop Products, Wilmington, DE, USA). cDNA was synthesized from 2 µg of RNA using the Superscript III® First-Strand Synthesis System (Invitrogen) following the manufacturer’s instructions.
Gene expression by quantitative PCR (RT-qPCR)
To determine the expression profiles of RmATG3, RmATG4, RmATG6, RmATG8a and RmATG8b genes in eggs and larvae of R. microplus, specific primers were designed (Table 1). PCR was initially standardized by final point PCR and observation in 1% agarose gels to validate the primers at an average Tm of 64 °C. β-actin (ACTB), elongation factor 1-α (ELFIA), ribosomal protein L4 (RPL4), acidic ribosomal protein P0 (P0) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) genes were used as reference genes (Nijhof et al. 2009; Umemiya-Shirafuji et al. 2014). RT-qPCR amplifications were performed as described previously (Flores et al. 2016) with some modifications. Briefly, amplifications were carried out in 96-well plates using SYBR Green detection chemistry in a StepOnePlus™ RT-qPCR System (Applied Biosystems/Ambion, Austin, TX, USA). Each single reaction (20 µl) contained 0.4 µl of first strand cDNA template (100 ng µl−1), 0.5 µl of forward and reverse primers (10 µM), 10 µl of SYBR Select Master Mix and 8.6 µl of sterile deionized water. Cycling conditions consisted of initial holding stage at 48 °C for 30 s, followed by denaturation step at 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s, 60 °C for 1 min and 72 °C for 30 s. Amplification was followed by melting curve analysis to confirm the specificity of the reactions. Baseline and quantification cycles (Cq) were automatically determined using RT-qPCR System software. PCR reactions were run in triplicate for each sample in three independent experiments. We routinely run no template controls (NTC) in all our PCR plates, as well as blank controls. Together with reference and testing genes for each plate. Relative expression was calculated using the comparative quantitative cycle method (Livak and Schmittgen 2001), where the endogenous reference gene ELFIA transcript level was utilized to normalize the expression for the different ATG genes, which turned out to be the most stably expressed control gene during the experiment.
Bioinformatic analysis
Primers were designed with the Primer3 software v.4.1.0 (http://primer3.ut.ee/; Untergasser et al. 2012). NormFinder algorithm was used to identify the optimal normalization gene among the set of reference genes (Andersen et al. 2004). Alignment and phylogenetic reconstructions were performed using “build” of ETE3 v.3.0.0b32 (Huerta-Cepas et al. 2016) as implemented on the GenomeNet (https://www.genome.jp/tools/ete/). Maximum Likelihood tree was inferred using RAxML v.8.1.20 ran with model GTRGAMMA and default parameters (Stamatakis 2014). Branch supports were computed out of 100 bootstrapped trees.
Statistical analysis
Expression data of RmATG3, RmATG4, RmATG6, RmATG8a and RmATG8b were analyzed by one-way ANOVA using Statgraphics™ Centurion XVI.I software. Tukey’s test was used to determine significant differences of each gene at different time points during development of R. microplus. PCR data were expressed as mean ± SEM in the method (2−∆∆CT).
Results and discussion
We determined the expression stability of the set of candidate normalization genes, during the experiment period, using the NormFinder algorithm (Andersen et al. 2004). According to their expression stability values, candidate genes were ranked as follows: ELFIA < ACTB < RPL4 < GAPDH < P0. The candidate gene most stably expressed (which had the smallest stability value) was ELFIA; therefore, it was used to normalize the expression of ATG genes.
Expression of ATG genes during embryogenesis of Rhipicephalus microplus
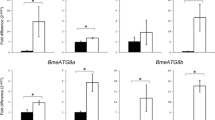
Life cycle in the one-host tick R. microplus occurs in two phases, being the first a free-living phase and the second a parasitic phase. This study focuses only on the first one. Once the eggs have been laid embryonic development commences. Under laboratory conditions this stage lasts approximately 24 days (Davey et al. 1980; Senbill et al. 2018). In our study, we observed that embryonic development lasted 23 days under our experimental conditions. We determined the expression profiles of ATG genes on egg samples collected every 5 days. Freshly laid eggs (H0 sample) showed low expression of all ATG genes but RmATG8b (Fig. 1). Umemiya-Shirafuji et al. (2014) reported that HlATG3, HlATG4, HlATG8 and HlATG12 genes were expressed at the highest level in developing eggs on day zero after oviposition in Haemaphysalis longicornis tick. These transcripts are most likely maternally inherited to the eggs. In sharp contrast, RmATG3, RmATG4, RmATG6 and RmATG8a mRNAs were significantly up-regulated on eggs at 5 days of embryonic development, whereas, RmATG8b mRNA showed a diminished expression. This up-regulation parallels the embryo cellularization, which characterize the cellular blastoderm stage (Campos et al. 2006). In Drosophila melanogaster embryos, a burst of spatially regulated autophagy during late cellularization has been observed, which is controlled via the autophagy-initiating kinase Atg1 and downstream Atg proteins (Kuhn et al. 2015). In both organisms, autophagy might participate in the degradation of maternal proteins during the maternal to zygotic transition and/or in supporting embryo development by supply of nutrients derived from cytosol, organelles, lipids and carbohydrates catabolism. In accordance with this hypothesis, it has been reported that the total lipid and carbohydrate content decreased between the 5th and 7th days and between the 7th and 9th days of R. microplus embryo development, respectively. Conversely, total RNA amount increased between the 6th and the 9th days after oviposition, indicating the activation of zygotic transcription (Campos et al. 2006). Following embryonic development, most ATG genes were down-regulated on H10 samples. However, a second wave of ATG genes up-regulation (all but RmATG8b) was clearly evident on H15 sample, whereas on H20 sample, RmATG3, RmATG8a and RmATG8b genes were also up-regulated (Fig. 1). These expression patterns concur with developmental changes in R. microplus tick embryos, such as fourth leg pair regression and its eventual disappearance, fading of the transient ventral furrow, opisthosomal cells dorsal migration and surrounding of the yolk mass and finally dorsal closure (Santos et al. 2013). With regard to energy sources, the end of embryogenesis is characterized by lower total lipid and sugar contents compared to early embryonic development (Campos et al. 2006). Moreover, it has been estimated that during R. microplus embryo development about 40% of vitellin, the major yolk protein, is consumed. Vitellin is intensely degraded during the first five days and in the last week of development, when organogenesis has been completed (Logullo et al. 2002; Campos et al. 2006). In H. longicornis tick, HlATG3, HlATG4 and HlATG12 genes were down-regulated before or at the onset of organogenesis. In contrast, HlATG8 gene was up-regulated from 10 days after oviposition and its expression was sustained until the end of embryogenesis. This augmented expression seemed to be concurrent with the onset of the differentiation of larval organs (Umemiya-Shirafuji et al. 2014). However, how autophagy participates in the energy metabolism and/or in the tick embryonic development at present is unknown. Evidence from other organisms suggests that autophagy may be essential in regulating both processes. For example, during Caenorhabditis elegans embryogenesis, maternally-loaded germline P granule components in somatic cells undergo selective autophagic degradation (Zhao et al. 2009). Moreover, autophagy-dependent degradation of ribosomal RNA is essential for maintaining nucleotide homeostasis during C. elegans development (Liu et al. 2018). In D. melanogaster, null mutants for diverse ATG genes are differentially affected on their development. For example, null mutants of Atg2 and Atg18 exhibit late pupal/pharate adult lethality. Similarly, mutants of Atg1, Atg13 and FIP200 die as pharate adults. Atg6, Vps34, Vps15 and UVRAG mutants die as L3 stage larvae. Interestingly, null mutants of Atg7, Atg8a and Atg16 are viable but exhibit a shorter lifespan and increased stress sensitivity (reviewed by Mulakkal et al. 2014).
Expression profiles of RmATG genes during embryogenesis and increasing larval starvation in Rhipicephalus microplus. aRmATG3; bRmATG4; cRmATG6; dRmATG8a; eRmATG8b. Total RNA was extracted from eggs (samples H0-H20) and larvae (samples L0-L50) at indicated times and used for the cDNA synthesis. The relative expression levels of RmATG genes were determined by RT-qPCR and normalized by the expression level of ELFIA. Data represent the mean ± SEM of triplicate samples from three independent biological replicates. *Significant up-regulation and # significant down-regulation (P < 0.05). Note the different scales on the y-axes
Expression of ATG genes under increasing larval starvation
In order to proceed to further developmental stages, newly hatched larvae must find a suitable host. However, the host-seeking period can last from a few days to several months. It is known that R. microplus larvae can survive up to 240 days without food, at 22 °C and 90% relative humidity (Hitchcock 1955). In the absence of exogenous nutrients, their survival depends exclusively on the remaining nutrients brought from the egg. As an example, it has been calculated that exhaustion of remaining vitellin (~ 60%) is reached at 40 days old larvae (Miranda–Miranda et al. 2009) Under such a scenario, we assessed the expression level of the ATG genes under study, on larvae collected every 5 days during 50 days (recreating an increasing starvation condition). Newly eclosioned R. microplus larvae (L0 sample) showed low expression of all ATG genes evaluated, excepting RmATG8a (Fig. 1). Low expression remained on L5 and L10 samples (5 and 10 days after eclosion, respectively). However, from L15 sample onwards, successive waves of transcriptional activation of ATG genes were observed. RmATG3 and RmATG6 genes were up-regulated on L15, L25 and L40 samples; whereas RmATG4 and RmATG8a genes were up-regulated on L15, L25, L40 and L45 samples. With regard to RmATG8b gene, this was up-regulated on L15, L40 and L45 samples. This cyclical pattern of positive gene regulation occurred roughly every 10–15 days, as starvation progressed. This transcriptional activation appears to be necessary for sustained autophagy, particularly to replenish consumables such as Atg8 proteins, which are degraded along with captured cargo in autophagolysosomes (Deretic et al. 2013). Recently, Rosendale et al. (2019) examined the bioenergetics, transcriptomic and behavioural changes of female American dog ticks, Dermacentor variabilis, over the course of prolonged starvation (up to 9 months post-blood meal). They found that several ATG genes were differentially expressed during starvation. Particularly, seven genes (ATG2A, ATG4B, ATG8b, ATG9A, ATG13, ATG14 and ATG16 X1) tended to increase their expression with starvation. In another study, a set of ATG genes (ulk1, lc3b, atg7 and atg12) was monitored during a long-term fasting (60 days) in European eel (Anguilla anguilla). Inspected genes displayed a non-random fluctuating expression pattern (Bolliet et al. 2017). Of note, the five ATG genes here studied are present in other ticks and in other arthropods. We performed a phylogenetic analysis including sequences of different ATG genes from several ticks (H. longicornis, R. microplus, Ixodes scapularis, D. variabilis) and insects (D. melanogaster, Apis mellifera) (Table 2). The phylogenetic tree showed that ATG sequences form distinctive clusters (e.g. ATG3, ATG4, ATG6, ATG8), suggesting that these genes are homologous among species (Fig. 2). Taken together, these results suggest that autophagy is playing a significant role in nutrient regulation during starvation. The observed cyclic alternating expression pattern of ATG genes, under increasing starvation, indicates that the genetic program of autophagy operates on demand, as nutrients are required to permit tick survival, in the absence of exogenous nutrients. The elucidation of mechanisms through which autophagy control nutrient availability and utilization is an active field of research.
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Bolliet V, Labonne J, Olazcuaga L, Panserat S, Seiliez I (2017) Modeling of autophagy-related gene expression dynamics during long term fasting in European eel (Anguilla anguilla). Sci Rep 7:17896
Campos E, Moraes J, Façanha AR, Moreira É, Valle D, Abreu L, Manso PPA, Nascimento A, Pelajo-Machado M, Lenzi H, Masuda A, da Silva IV Jr., Logullo C (2006) Kinetics of energy source utilization in Boophilus microplus (Canestrini, 1887) (Acari: Ixodidae) embryonic development. Vet Parasitol 138:349–357
Davey RB, Garza J Jr, Thompson GD, Drummond RO (1980) Ovipositional biology of the southern cattle tick, Boophilus microplus (Acari: Ixodidae), in the laboratory. J Med Entomol 17(2):117–121
Deretic V, Saitoh T, Akira S (2013) Autophagy in infection, inflammation and immunity. Nat Rev Immunol 13:722–737
Flores FJM, Gutiérrez OA, Rosario CR, Padilla CE, Álvarez AH, Martínez VM (2014) Molecular cloning and characterization of two novel autophagy-related genes belonging to the ATG8 family from the cattle tick Rhipicephalus (Boophilus) microplus (Acari: Ixodidae). Exp Appl Acarol 64(4):533–542
Flores FJM, Barragán ACP, Sánchez HCV, Padilla CE, González CC, Ortuño SD, Martínez VM (2016) Molecular characterization and expression analysis of three novel autophagy-related genes from the cattle tick Rhipicephalus (Boophilus) microplus (Acari: Ixodidae). Parasitology 143:1802–1809
Hitchcock LF (1955) Studies of the non-parasitic stages on the cattle tick, Boophilus microplus (Canestrini) (Acarina: Ixodidae). Aust J Zool 3(2):145–155
Huerta-Cepas J, Serra F, Bork P (2016) ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 33:1635–1638
Kuhn H, Sopko R, Coughlin M, Perrimon N, Mitchinson T (2015) The Atg1-Tor pathway regulates yolk catabolism in Drosophila embryos. Development 142:3869–3878
Liu Y, Zou W, Yang P, Wang L, Ma Y, Zhang H, Wang X (2018) Autophagy-dependent ribosomal RNA degradation is essential for maintaining nucleotide homeostasis during C. elegans development. eLife 7:e36588
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25:402–408
Logullo C, Moraes J, Dansa-Petretski M, JrIS Vaz, Masuda A, Sorgine MHF, Braz GR, Masuda H, Oliveira PL (2002) Binding and storage of heme by vitellin from the cattle tick, Boophilus microplus. Insect Biochem Mol Biol 32:1805–1811
Miranda-Miranda E, Cossio-Bayugar R, Quezada-Delgado MDR, Olvera-Valencia F, Neri-Orantes S (2009) Age-induced carboxylesterase expression in acaricide-resistant Rhipicephalus microplus. Res J Parasitol 4(3):70–78
Mulakkal NC, Nagy P, Takats S, Tusco R, Juhász G, Nezis IP (2014) Autophagy in Drosophila: from historical studies to current knowledge. BioMed Res Int. https://doi.org/10.1155/2014/273473
Nijhof AM, Balk JA, Postigo M, Jongejan F (2009) Selection of reference genes for quantitative RT-PCR studies in Rhipicephalus (Boophilus) microplus and Rhipicephalus appendiculatus ticks and determination of the expression profile of Bm86. BMC Mol Biol 10:112
Nuñez JL, Muñoz-Cobeñas ME, Moltedo HL (1985) Boophilus microplus. The common cattle tick. Springer, Berlin, p 204
Rosendale AJ, Dunlevy ME, McCue MD, Benoit JB (2019) Progressive behavioural, physiological and transcriptomic shifts over the course of prolonged starvation in ticks. Mol Ecol 28:49–65
Santos VT, Ribeiro L, Fraga A, de Barros CM, Campos E, Moraes J, Fontenele MR, Araújo HM, Feitosa NM, Logullo C, da Fonseca RN (2013) The embryogenesis of the tick Rhipicephalus (Boophilus) microplus: the establishment of a new chelicerate model system. Genesis 51:803–818
Senbill H, Hazarika LK, Baruah A, Borah DK, Bhattacharyya B, Rahman S (2018) Life cycle of the southern cattle tick, Rhipicephalus (Boophilus) microplus Canestrini 1888 (Acari: Ixodidae) under laboratory conditions. Syst Appl Acarol 23(6):1169–1179
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Umemiya-Shirafuji R, Galay RL, Maeda H, Kawano S, Tanaka T, Fukumoto S, Suzuki H, Tsuji N, Fujisaki K (2014) Expression analysis of autophagy-related genes in the hard tick Haemaphysalis longicornis. Vet Parasitol 201:169–175
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3-new capabilities and interfaces. Nucleic Acids Res 40(15):e115
Zhao Y, Tian E, Zhang H (2009) Selective autophagic degradation of maternally-loaded germline P granule components in somatic cells during C. elegans embryogenesis. Autophagy 5(5):717–719
Acknowledgements
This work was supported by Consejo Nacional de Ciencia y Tecnología (CONACYT) México; Grant Numbers 216321 and 246310, both to MMV.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
González Castillo, C., Ortuño Sahagún, D. & Martínez Velázquez, M. Expression dynamics of autophagy-related genes in the cattle tick Rhipicephalus microplus during embryonic development and under increasing larval starvation. Exp Appl Acarol 79, 255–266 (2019). https://doi.org/10.1007/s10493-019-00428-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-019-00428-7