Abstract
The aim of this study was to investigate the genetic diversity of Varroa destructor parasitizing Apis mellifera colonies and to test for possible host–parasite association at the mitochondrial DNA (mtDNA) level. Six A. mellifera haplotypes (including a novel C2aa) and five haplotypes of V. destructor were detected in 29 analyzed colonies from eight sampling sites in Serbia. We revealed the presence of the K and S1 haplotypes as well as KS1 and KP1 heteroplasmic mite individuals in all localities, while the P1 haplotype was only found in four sampling sites. Significant differences in V. destructor genetic diversity were found at both apiary and colony levels, with mite haplotypes coexisting in almost all tested colonies. In addition, a significant correlation between the number of analyzed mites per colony and the number of identified V. destructor haplotypes was observed. However, no significant host–parasite relationship was found, suggesting that mites bearing different haplotypes as well as those heteroplasmic individuals are well adapted to the host, A. mellifera, independently of the identified haplotype present in each colony. Our results will contribute to future population and biogeographic studies concerning V. destructor infesting A. mellifera, as well as to better understanding their host–parasite relationship.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The honey bee (Apis mellifera L.), is considered the most important pollinator of crop monocultures worldwide (De la Rúa et al. 2009). Because of its pollination success and honey production, A. mellifera has been spread and repeatedly introduced in non-endemic regions, including eastern Asia (Moritz et al. 2005; Pirk et al. 2017). In this context, beekeeping management brought A. mellifera into contact with indigenous Asian honey bee species and their parasitic mites (Anderson and Morgan 2007), including Varroa destructor Anderson and Trueman and microsporidian parasites (Higes et al. 2006).
Varroa destructor is an obligate ectoparasitic mite, feeding on the fat body tissue of honey bees (Ramsey et al. 2019) and vectoring multiple viruses (Genersch and Aubert 2010). After it host shifted from the Asian honey bee Apis cerana Fabricius, it has spread globally (Anderson and Trueman 2000; Rosenkranz et al. 2010), significantly contributing to colony losses of A. mellifera worldwide (vanEngelsdorp et al. 2008; Genersch et al. 2010). Initially, only two mitochondrial DNA (mtDNA) haplotypes of V. destructor (Korea and Japan, K and J haplotype, respectively) successfully colonized A. mellifera outside Asia (Anderson and Trueman 2000). Although a high mtDNA diversity of V. destructor was detected primarily on A. cerana, four haplotypes parasitizing A. mellifera throughout Asia were described using more sensitive mtDNA markers (Navajas et al. 2010). Recent studies have reported well-established infestations of A. mellifera by related Luzon 1 mite haplotype and sibling Varroa jacobsoni Oudemans species, revealing new host shift events taking place in Southeast Asia and Oceania, respectively (Beaurepaire et al. 2015; Roberts et al. 2015).
Varroa destructor was detected in Serbia for the first time in 1976 (Lolin 1977) and mites originating from this region (former Yugoslavia) were later all identified as bearing the K haplotype as defined by Anderson and Trueman (2000). However, novel Serbia 1 (S1) and Peshter 1 (P1) haplotypes of V. destructor from Serbia were described in 2013, differing from the original K haplotype in single nucleotide polymorphisms (SNPs) within cytochrome c oxidase 1 (cox1) and cytochrome b (cytb) gene sequences, respectively (Gajić et al. 2013). Lately, nucleotide heteroplasmy has been discovered in this parasitic species and a molecular assay for identification of defined haplotypes and heteroplasmic individuals has been established (Gajić et al. 2016).
Biogeographic studies based on molecular analyses of the host species A. mellifera in Balkan countries revealed that two subspecies, A. m. carnica and A. m. macedonica, are present in Serbia (Stevanovic et al. 2010; Nedić et al. 2014, respectively). In total seven haplotypes were detected, all belonging to the East-Mediterranean (C) evolutionary lineage of honey bee subspecies, among them two newly-described, C2o and C2p (Muñoz et al. 2012). These findings are in agreement with a status of Balkan Peninsula as a centre of insect biodiversity (Milankov et al. 2009) with a central position of Serbia in A. mellifera C-lineage distribution area (Meixner et al. 2013).
Having in mind observed mitochondrial diversity of both A. mellifera and V. destructor in Serbia (Stevanovic et al. 2010; Muñoz et al. 2012; Gajić et al. 2013, 2016), we hypothesized that the distribution of existing V. destructor haplotypes and heteroplasmic mites depends on the host haplotype. Therefore, the aim of this study was to investigate the genetic diversity and distribution of V. destructor in genetically distinct A. mellifera colonies from Serbia and to examine possible association of the host and parasite based on mtDNA variation.
Materials and methods
Honey bee and mite samples
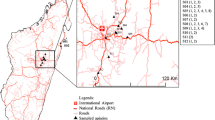
Honey bee workers and V. destructor females were collected in 29 A. mellifera colonies from eight localities throughout Serbia in 2012. Samples were taken from a single apiary at each locality (Fig. 1a). Colonies from five localities were stationary (Belgrade, Zlatibor, Niš, Suvi Do, Šaprance) whereas those from the remaining three localities (Palić, Lapovo, Boljevac) were migratory. All honey bee and mite samples were transported to the laboratory in 70% ethanol and stored at − 20 °C until DNA extraction. One honey bee worker per colony (n = 29) and 2–14 mites per colony (n = 245) were used for molecular analyses (Table 1).
DNA extraction
Total DNA was extracted from whole adult V. destructor females and from three right legs of one worker honey bee per colony (Evans et al. 2013). Individual mites were previously washed in distilled H2O, dried on filter paper and crushed with sterile mini-pestles in 1.5-mL plastic tubes. DNA was then extracted using KAPA Express Extract Kit (KAPA Biosystems, South Africa), according to the manufacturer’s protocol.
Molecular identification
Varroa destructor
Initial typing of all 245 Varroa mites was based on the presence/absence of SNPs and heteroplasmy using amplification refractory mutation system (ARMS) in partial cox1 sequence and PCR-RFLP in cytb gene, according to the detailed protocols described elsewhere (Gajić et al. 2016). Final detection of one of the previously identified mtDNA haplotypes (K, S1, P1) or heteroplasmic V. destructor individuals (KS1, KP1) was done by coupling results obtained for individual cox1 and cytb sequences.
Apis mellifera
For haplotype identification, the tRNAleu-cox2 intergenic region of A. mellifera was amplified using KAPA Taq PCR Kit (KAPA Biosystems, South Africa). Reaction volume of 25 µL consisted of 14.4 µL nuclease free water, 2.5 µL 10× KAPA Taq buffer, 0.5 µL (10 mM) of dNTP mix, 1.25 µL (10 µM) of primers E2 and H2 (Garnery et al. 1991), 0.1 µL (5U/µL) of KAPA Taq DNA polymerase and 5 µL of DNA template. Amplification protocol comprised initial denaturation at 95 °C for 5 min, 36 cycles at 95 °C for 45 s, annealing at 47 °C for 60 s and extension at 72 °C for 90 s, followed by final extension at 72 °C for 10 min (Evans et al. 2013).
Sequencing
Mitochondrial amplicons of cox1 and cytb fragments of two mites per colony (n = 58), also analyzed by ARMS and PCR-RFLP assays, were Sanger sequenced using the same primer sets as for PCR amplification to confirm accuracy of Varroa genotyping method. Amplicons of the tRNAleu-cox2 intergenic region of one honey bee per colony (n = 29) were also Sanger sequenced with the primers used for PCR amplification. Bioinformatic analysis of resulting sequences was done using the software BioEdit v. 7.1.3 (Hall 1999).
Statistical analysis
The χ2 test was used to compare: (1) the distribution of V. destructor haplotypes depending on locality, (2) the distribution of V. destructor haplotypes depending on beekeeping practice, and (3) the frequency of SNP and heteroplasmy depending on the analyzed sequence. General discriminant analysis (squared Mahalanobis’ distance and its significance) determined the significance of differences in distribution of V. destructor haplotypes between the localities, between the hives, and between various bee haplotypes. The hierarchical model of the cluster analysis, based on the squared Mahalanobis’ distances and the Ward’s method of grouping, provided a dendrogram, pointing to groups of similar elements. Spearman's rank correlation coefficient quantified the dependence between the number of analyzed mites per colony and the number of identified V. destructor mtDNA haplotypes. Statistical analysis of experimental data was performed using software package STATISTICA v. 7.0 (Statsoft, Tulsa, OK, USA).
Results
Varroa destructor haplotypes
Independent analyses of cox1 and cytb sequences
In 48.2% (118/245) of mite samples, cox1 sequence was identical to the reference sequence of the K haplotype (GenBank Accession No. GQ379056), showing no SNPs or heteroplasmy. Cytb sequence corresponding to the K haplotype (GenBank Accession No. GQ379094) was observed in 83.7% (205/245) of mites. However, SNPs or heteroplasmy was observed in 51.8 and 16.3% of mites in cox1 and cytb, respectively. Moreover, the occurrence of SNPs and heteroplasmy was significantly dependent on the analyzed sequence (χ2 = 67.190, df = 1, p < 0.001). All sequencing results for 58 mites were in agreement with results obtained using ARMS and PCR-RFLP methods for cox1 and cytb sequence, respectively.
Concatenated sequence analysis
Based on concatenated cox1 and cytb sequence analysis, the majority of mite samples belonged to either K or S1 haplotypes (31.8% of each). Cox1 heteroplasmy (KS1 individuals) was found in 20% of cases, cytb heteroplasmy (KP1 individuals) was observed in 11.8%, whereas 4.5% of mites were assigned to the P1 haplotype. There were no samples with simultaneously detected SNPs or heteroplasmy in both (cox1 and cytb) analyzed sequences, neither individuals containing SNP in one sequence and heteroplasmy within the other (hypothetical S1-P1, S1-KP1, KS1-P1 and KS1-KP1 individuals) (Table 1).
Distribution of Varroa destructor mtDNA haplotypes
Distribution among localities
The K and S1 haplotypes as well as KS1 and KP1 heteroplasmic individuals were registered at all sampling sites. However, P1 haplotype was found only in four localities with stationary beekeeping practice (Belgrade, Zlatibor, Suvi Do and Šaprance) (Table 1). Moreover, the frequency of detected V. destructor mtDNA haplotypes was highly dependent on the practice performed by beekeepers that is migratory or stationary beekeeping (χ2 = 14.258, df = 4, p = 0.007).
At most sites, predominant haplotypes were K or S1, with KP1 individuals as the most frequent in Boljevac (Table 1). Genetic diversity of V. destructor populations was significantly different among localities observed simultaneously (χ2 = 78.392, df = 28, p < 0.001). In addition, significant differences were also detected among individual localities, showing different haplotype frequency of mites from Zlatibor compared to those from all other localities (p < 0.001) except from Šaprance.
Hierarchical clustering of localities according to the similarity of mite haplotypes using squared Mahalanobis’ distance between localities and Ward’s method of grouping, showed three clusters: (1) Boljevac, Niš, Palić; (2) Šaprance, Lapovo, Suvi Do, Belgrade; (3) Zlatibor (Fig. 1b).
Distribution among and within honey bee colonies
Most hives at the same apiary had similar distribution of mite haplotypes (Online Resource 1). However, based on the levels of squared Mahalanobis’ distance, hives containing mites with P1 haplotype in Belgrade and Zlatibor were significantly different from their neighboring colonies (p < 0.05).
Number of identified mite mtDNA variants per hive ranged from 1 to 5, with 6.9% (2/29) colonies being infested by only S1 haplotype and with coexistence of mite haplotypes registered in 93.1% (27/29) colonies (Table 2). Spearman's rank correlation coefficient (ρ = 0.711, p < 0.001) showed strong dependence between the number of analyzed mites per colony and the number of identified V. destructor mtDNA haplotypes.
Apis mellifera haplotypes
Based on the nucleotide polymorphism detected in the 571–573 bp long tRNAleu-cox2 sequences, six A. mellifera haplotypes were identified, five of them previously described (C1a, C2d, C2e, C2i, C2c; GenBank Accession Nos. JQ977699–JQ977703). One sequence characterized by the presence of a new SNP was identified for the first time in this study and assigned to the novel C2aa haplotype (GenBank Accession No. MG788257) following the nomenclature of Rortais et al. (2011) and Chávez-Galarza et al. (2017). Number of detected honey bee haplotypes per locality ranged from one to three. The most frequent haplotype was C2d, followed by C2e, C1a and C2c, with the lowest frequency registered for C2i and C2aa haplotypes (Table 1).
Host–parasite association
Mites with K and S1 haplotypes were found in different proportion on honey bee colonies bearing all identified haplotypes, whereas mites with P1 haplotype were registered only in colonies with C2d and C2e haplotypes. KS1 and KP1 heteroplasmic individuals parasitized honey bees with C1a, C2d, C2e and C2c haplotypes, whereas mite KS1 individuals was also found in honey bees bearing with C2aa (Fig. 2). Significance levels of squared Mahalanobis’ distances showed that genetic structure of V. destructor population was not dependent on the host haplotype (p > 0.05).
However, distribution of V. destructor haplotypes and heteroplasmic individuals infesting honey bee bearing C2d haplotype in Boljevac and Zlatibor was significantly different from mite distribution in honey bees with C2d haplotype in the remaining localities (p ≤ 0.031 and p ≤ 0.002, respectively), as well as between Belgrade and Palić (p = 0.029). Moreover, distribution of V. destructor mtDNA variants parasitizing C2e honey bees in Zlatibor significantly differed from all other localities (p ≤ 0.005).
Discussion
In the current study, we revealed that different V. destructor haplotypes and heteroplasmic individuals are widely distributed and coexist in A. mellifera colonies throughout Serbia, with no strict host–parasite association at mtDNA level.
Based on independent sequence analysis, significantly more V. destructor samples from our study showed identity to the reference K haplotype in cytb compared to the cox1 sequence (83.7 and 48.2%, respectively). However, Navajas et al. (2010) failed to detect any mutation in the cytb within the K haplogroup of V. destructor parasitizing A. mellifera in Asia. Additionally, in mites with SNPs or heteroplasmy detected in one sequence (cox1 or cytb), the second analyzed sequence was always without observed SNP and heteroplasmy being therefore identical to the K haplotype. This SNP-heteroplasmy pattern put forward that point mutations in cox1 and cytb occurred independently as well as in different mite populations of the original K haplotype.
Based on concatenated cox1 and cytb sequence analysis, the K and S1 haplotypes were equally represented (each by 31.8%) in tested samples whereas the P1 haplotype was detected in only 4.5% of mites. All previous studies showed predominance of the K haplotype outside Asia (Anderson and Trueman 2000; Garrido et al. 2003; Solignac et al. 2005; Muñoz et al. 2008; Farjamfar et al. 2018) including the geographic region of former Yugoslavia (Anderson and Trueman 2000). Surprisingly low prevalence of the K haplotype (in less than one third of analyzed samples) observed in the current study can be explained by better adaptation or higher virulence of coexisting haplotypes and heteroplasmic mites. Moreover, temporal changes in V. destructor haplotype are not unusual, as it has been reported for the K haplotype displacing the J haplotype in Brazil (Garrido et al. 2003).
Heteroplasmic KS1 and KP1 individuals were found in 20.0 and 11.8% of samples, respectively, suggesting that heteroplasmy is common in V. destructor populations. In contrast to the high frequency of heteroplasmic individuals in all investigated localities in Serbia, heteroplasmy has not been reported in previous mtDNA-based studies concerning Varroa spp. regardless the number and length of analyzed mitochondrial fragments (Solignac et al. 2005; Navajas et al. 2010). In addition, site heteroplasmy in V. destructor has not been detected independently of whether DNA was extracted from individual mites (Zhou et al. 2004; Solignac et al. 2005; Muñoz et al. 2008; Navajas et al. 2010) or from a pooled mite sample (Maggi et al. 2012). Moreover, the absence of S1-P1, S1-KP1, KS1-P1 and KS1-KP1 Varroa haplotypes in this study emphasizes maternal inheritance of mtDNA in V. destructor and minimizes the role of paternal leakage as the cause of heteroplasmy in this species. Nevertheless, additional analyses are needed to elucidate the source and transmission mechanism of the observed heteroplasmy.
We found that coexistence of V. destructor haplotypes and heteroplasmic individuals within the same colony is widely present. This finding enables monitoring of seasonal dynamics of coexisting haplotypes and their cumulative effect on honey bee colony, but making at the same time the virulence estimation of particular mite haplotypes more challenging. Additionally, a high correlation observed between the number of the analyzed mites per colony and the number of identified mtDNA variants of V. destructor needs future studies in order to define the optimal number of mite samples for reliable molecular characterization.
A high mitochondrial diversity is present in A. mellifera colonies located in Serbia where eight haplotypes have been found, in contrast to neighboring countries where one (in Slovenia and Macedonia), two (in Albania) and four (in Croatia) haplotypes were detected (Sušnik et al. 2004; Muñoz et al. 2009, 2012; Stevanovic et al. 2010). This high genetic diversity can be explained by the ecological history of the Balkan Peninsula that was one of the major Pleistocene ice age refugia for various species and consequently, the source for rapid post-glacial colonization of Europe (Hewitt et al. 1999).
A novel bee haplotype (C2aa) was identified in Syenichko-Peshterski region, where the micro-endemic, unique C2p haplotype was previously found (Muñoz et al. 2012). Importantly, C2aa, the only newly-found haplotype in this study, as well as C2p from previous study (Muñoz et al. 2012) originated from stationary apiaries situated at the altitude over 1000 m. The presence of those two haplotypes could be due to adaptation to specific ecological conditions as observed in the Carpathians (Coroian et al. 2014) if the beekeepers maintain their queens exclusively.
Our survey did not reveal any significant host–parasite haplotype correlation, despite mtDNA variation of A. mellifera and V. destructor in Serbia. These results are in line with those of Muñoz et al. (2008), who failed to detect significant relationship in the host–parasite haplotype distribution between A. m. iberiensis and V. destructor in an apiary in central Spain. However, we found that the frequency of observed V. destructor haplotypes was significantly dependent on the performed beekeeping practice. This finding indicates that migratory hives could facilitate dispersion of mites via horizontal transmission and lead to homogenization of V. destructor genetic structure, as it was reported at the apiary level (Beaurepaire et al. 2017; Dynes et al. 2017).
The lack of host specificity of Luzon 1 haplotype to both A. mellifera and A. cerana (Beaurepaire et al. 2015), together with the finding of two genetic haplotypes of the sister species V. jacobsoni reproducing on A. mellifera (Roberts et al. 2015) suggest the ongoing evolution and further host switching of Varroa spp. in Asia. However, coexisting haplotypes and heteroplasmic mites we detected in our study are apparently mutated descendants of the original K haplotype with the current host specificity to A. mellifera outside Asia, whose spreading potential should be carefully considered in future studies.
Conclusions
The coexistence of mtDNA haplotypes of V. destructor in A. mellifera colonies throughout Serbia emphasizes the need of analyzing more than one mite per colony for objective mite molecular typing and will make the virulence estimation of particular mite strain more challenging. In addition, high proportion of heteroplasmic mites detected in analyzed populations and specific reproductive behavior of V. destructor make this parasitic species a suitable model organism for any analysis concerning the cause, transmission and maintenance of heteroplasmy. Our findings will also contribute to future population and biogeographic studies concerning V. destructor infesting A. mellifera, as well as to better understanding their host–parasite relationship in other European countries.
References
Anderson DL, Morgan MJ (2007) Genetic and morphological variation of bee-parasitic Tropilaelaps mites (Acari: Laelapidae): new and re-defined species. Exp Appl Acarol 43(1):1–24
Anderson DL, Trueman JWH (2000) Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp Appl Acarol 24(3):165–189
Beaurepaire AL, Ellis JD, Krieger KJ, Moritz RF (2017) Association of Varroa destructor females in multiply infested cells of the honeybee Apis mellifera. Insect Sci. https://doi.org/10.1111/1744-7917.12529
Beaurepaire AL, Truong TA, Fajardo AC, Dinh TQ, Cervancia C, Moritz RF (2015) Host specificity in the honeybee parasitic mite, Varroa spp. in Apis mellifera and Apis cerana. PLoS ONE 10(8):e0135103
Chávez-Galarza J, Garnery L, Henriques D, Neves CJ, Loucif-Ayad W, Jonhston JS, Pinto MA (2017) Mitochondrial DNA variation of Apis mellifera iberiensis: further insights from a large-scale study using sequence data of the tRNA leu-cox2 intergenic region. Apidologie 48(4):533–544
Coroian CO, Muñoz I, Schlüns EA, Paniti-Teleky OR, Erler S, Furdui EM, Mărghitaş LA, Dezmirean DS, Schlüns H, De la Rúa P, Moritz RFA (2014) Climate rather than geography separates two European honeybee subspecies. Mol Ecol 23(9):2353–2361
De la Rúa P, Jaffé R, Dall'Olio R, Muñoz I, Serrano J (2009) Biodiversity, conservation and current threats to European honeybees. Apidologie 40(3):263–284
Dynes TL, De Roode JC, Lyons JI, Berry JA, Delaplane KS, Brosi BJ (2017) Fine scale population genetic structure of Varroa destructor, an ectoparasitic mite of the honey bee (Apis mellifera). Apidologie 48(1):93–101
Evans JD, Schwarz RS, Chen YP, Budge G, Cornman RS et al (2013) Standard methods for molecular research in Apis mellifera. J Apic Res 52(4):1–54
Farjamfar M, Saboori A, González-Cabrera J, Rodríguez CSH (2018) Genetic variability and pyrethroid susceptibility of the parasitic honey bee mite Varroa destructor (Acari: Varroidae) in Iran. Exp Appl Acarol 76(1):139–148
Gajić B, Radulovic Z, Stevanovic J, Kulisic Z, Vucicevic M, Simeunovic P, Stanimirovic Z (2013) Variability of the honey bee mite Varroa destructor in Serbia, based on mtDNA analysis. Exp Appl Acarol 61(1):97–105
Gajić B, Stevanović J, Radulović Ž, Kulišić Z, Vejnović B, Glavinić U, Stanimirović Z (2016) Haplotype identification and detection of mitochondrial DNA heteroplasmy in Varroa destructor mites using ARMS and PCR–RFLP methods. Exp Appl Acarol 70(3):287–297
Garnery L, Vautrin D, Cornuet JM, Solignac M (1991) Phylogenetic relationships in the genus Apis inferred from mitochondrial DNA sequence data. Apidologie 22(1):87–92
Garrido C, Rosenkranz P, Paxton RJ, Gonçalves LS (2003) Temporal changes in Varroa destructor fertility and haplotype in Brazil. Apidologie 34(6):535–541
Genersch E, Aubert M (2010) Emerging and re-emerging viruses of the honey bee (Apis mellifera L.). Vet Res 41(6):54. https://doi.org/10.1051/vetres/2010027
Genersch E, Von der Ohe W, Kaatz H, Schroeder A, Otten C et al (2010) The German bee monitoring project: a long term study to understand periodically high winter losses of honey bee colonies. Apidologie 41(3):332–352
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp 41:95–98
Hewitt GM (1999) Post-glacial re-colonization of European biota. Biol J Linn Soc 68(1–2):87–112
Higes M, Martín R, Meana A (2006) Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J Invertebr Pathol 92(2):93–95
Lolin M (1977) First case of varoatosis in our country. Pcelar 1–2:14–15 [in Serbian]
Maggi M, Medici S, Quintana S, Ruffinengo S, Marcángeli J, Martinez PG, Fuselli S, Eguaras M (2012) Genetic structure of Varroa destructor populations infesting Apis mellifera colonies in Argentina. Exp Appl Acarol 56(4):309–318
Meixner MD, Pinto MA, Bouga M, Kryger P, Ivanova E, Fuchs S (2013) Standard methods for characterising subspecies and ecotypes of Apis mellifera. J Apic Res 52(4):1–28
Milankov V, Ludoški J, Stahls G, Stamenković J, Vujić A (2009) High molecular and phenotypic diversity in the Merodon avidus complex (Diptera, Syrphidae): cryptic speciation in a diverse insect taxon. Zool J Linn Soc 155(4):819–833
Moritz RF, Härtel S, Neumann P (2005) Global invasions of the western honeybee (Apis mellifera) and the consequences for biodiversity. Ecoscience 12(3):289–301
Muñoz I, Dall’Olio R, Lodesani M, De la Rúa P (2009) Population genetic structure of coastal Croatian honeybees (Apis mellifera carnica). Apidologie 40(6):617–626
Muñoz I, Garrido-Bailón E, Martín-Hernández R, Meana A, Higes M, De la Rúa P (2008) Genetic profile of Varroa destructor infesting Apis mellifera iberiensis colonies. J Apic Res 47(4):310–313
Muñoz I, Stevanovic J, Stanimirovic Z, De la Rúa P (2012) Genetic variation of Apis mellifera from Serbia inferred from mitochondrial analysis. J Apic Sci 56(1):59–69
Navajas M, Anderson DL, De Guzman LI, Huang ZY, Clement J, Zhou T, Le Conte Y (2010) New Asian types of Varroa destructor: a potential new threat for world apiculture. Apidologie 41(2):181–193
Nedić N, Francis RM, Stanisavljević L, Pihler I, Kezić N, Bendixen C, Kryger P (2014) Detecting population admixture in honey bees of Serbia. J Apic Res 53(2):303–313
Pirk CW, Crewe RM, Moritz RF (2017) Risks and benefits of the biological interface between managed and wild bee pollinators. Funct Ecol 31(1):47–55
Ramsey SD, Ochoa R, Bauchan G, Gulbronson C, Mowery JD, Cohen A, Lim D, Joklik J, Cicero JM, Ellis JD, Hawthorne D, van Engelsdorp D (2019) Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc Natl Acad Sci USA 116(5):1792–1801
Roberts JMK, Anderson DL, Tay WT (2015) Multiple host shifts by the emerging honeybee parasite, Varroa jacobsoni. Mol Ecol 24(10):2379–2391
Rortais A, Arnold G, Alburaki M, Legout H, Garnery L (2011) Review of the DraI COI-COII test for the conservation of the black honeybee (Apis mellifera mellifera). Cons Gen Res 3(2):383–391
Rosenkranz P, Aumeier P, Ziegelmann B (2010) Biology and control of Varroa destructor. J Invertebr Pathol 103:S96–S119
Solignac M, Cornuet JM, Vautrin D, Le Conte Y, Anderson D, Evans J, Cros-Arteil S, Navajas M (2005) The invasive Korea and Japan types of Varroa destructor, ectoparasitic mites of the Western honeybee (Apis mellifera), are two partly isolated clones. Proc R Soc Lond B Biol 272(1561):411–419
Stevanovic J, Stanimirovic Z, Radakovic M, Kovacevic SR (2010) Biogeographic study of the honey bee (Apis mellifera L.) from Serbia, Bosnia and Herzegovina and Republic of Macedonia based on mitochondrial DNA analyses. Rus J Genet 46(5):603–609
Sušnik S, Kozmus P, Poklukar J, Meglic V (2004) Molecular characterisation of indigenous Apis mellifera carnica in Slovenia. Apidologie 35(6):623–636
vanEngelsdorp D, Hayes J, Underwood RM, Pettis J (2008) A survey of honey bee colony losses in the US, fall 2007 to spring 2008. PLoS ONE 3(12):e4071
Zhou T, Anderson DL, Huang ZY, Huang S, Yao J, Ken T, Zhang Q (2004) Identification of Varroa mites (Acari: Varroidae) infesting Apis cerana and Apis mellifera in China. Apidologie 35(6):645–654
Funding
This study was supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Grant No. III46002) and the 19908-GERM-15 project of Regional Excellence from the Fundación Séneca (Gobierno Regional de Murcia, Spain). Irene Muñoz is supported by Fundación Séneca (Gobierno Regional de Murcia, Spain) through the post-doctoral fellowship “Saavedra Fajardo” (20036/SF/16).
Author information
Authors and Affiliations
Contributions
BG designed the study and wrote the manuscript; BG, IM and PDLR conducted molecular analyses and bioinformatics; NL performed statistical analysis; JS, ZK and ZS helped in interpretation of obtained results and critically revised the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gajić, B., Muñoz, I., De la Rúa, P. et al. Coexistence of genetically different Varroa destructor in Apis mellifera colonies. Exp Appl Acarol 78, 315–326 (2019). https://doi.org/10.1007/s10493-019-00395-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-019-00395-z