Abstract
Segmentation is an essential task in modern medical imaging analysis. Since the scarcity of labeled pixel-level annotations often limits its wide applications, recent studies have proposed Semi-supervised Learning (SSL) frameworks to tackle this issue. Among them, the paradigm of pseudo-labeling, derived from SSL of natural images, has been popularly transferred on various medical datasets. Despite its promising results, we observe that many medical images’ regions are ambiguous, where pixels are challenging to be categorized as a specific class compared to natural images. Constructing hard pseudo-labels for these regions is consequently unintuitive and prone to be of low quality. To this end, we develop a novel SSL framework with the proposed Soft Pseudo-label Fusion strategy (called ”SPFSeg”). It can produce refined soft pseudo-labels, harboring the association knowledge between regions of interest (ROIs) and backgrounds while preserving the ”low-density” assumption of vanilla pseudo-labeling. These soft pseudo-labels can further establish potent supervision signals for unlabeled images, helping the segmentation model learn better feature representations. Through extensive experiments conducted on various datasets to evaluate the effectiveness of SPFSeg, our results manifest that its performance can surpass previous state-of-the-art semi-supervised frameworks on CXR-2014, ISIC-2017, and BUL-2020.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Medical imaging has widely been applied to modern medical analysis. Thereinto, segmentation is one of the most fundamental analysis tasks, assisting clinicians in focusing on pathological regions. Recently, with advances in Deep Learning, many researchers have attempted to apply Convolutional Neural Networks (CNNs) to realize automatic computer-aided segmentation tasks. Although they have demonstrated encouraging results on various datasets (e.g., skin lesion segmentation [1], lung segmentation [2], and cardiac MR image segmentation [3]), an inevitable challenge is that Deep Learning models require annotation-rich medical imaging datasets to guarantee their robustness and generalizability. In real scenarios, annotating medical images is particularly expensive and time-consuming because it needs to be conducted by experienced experts, and some images are hard to annotate due to their imaging qualities or scanning characteristics. This challenge is not beneficial to introducing these Deep Learning models into practical applications.
To this end, many studies have introduced Semi-supervised Learning (SSL) paradigms previously used in natural images to put forward some semi-supervised medical segmentation frameworks, seeking the possibility of leveraging unlabeled images when the number of labeled images is limited. Among them, Pseudo-labeling [4] has been extensively studied in medical imaging segmentation. Its main idea is to generate pixel-level pseudo-labels of unlabeled images and then use them to retrain the segmentation model in the same way as the labeled images. However, this paradigm often leads to low-quality pseudo-labels due to a lack of supervision from labeled images. Therefore, many studies [5,6,7] tend to refine them based on the properties of medical images. Although these frameworks significantly enhance the segmentation performance compared to the vanilla Pseudo-labeling strategy, an image property gap between natural and medical images may hinder the further improvement of segmentation performance. Specifically, as depicted in Fig. 1, many pixels in medical images’ regions, especially in the boundaries, are usually ambiguous and hard to be identified as specific categories compared to natural images. Therefore, it is unintuitive to produce hard (one-hot) pseudo-labels, as conducted in many existing pseudo-labeling-based studies.
The data samples of ISIC-2017, BUL-2020, and PASCAL VOC 2012. The “blue” shaded areas in medical images (ISIC-2017 and BUL-2020) are the ROIs annotated by human experts. The “yellow” boxes in medical images are the ambiguous boundary regions where the pixels are hard to be identified as a specific category. Conversely, the ROIs and backgrounds of natural images are relatively distinguishable
In knowledge distillation (see review in [8]), many response-based studies utilize the output soft targets from the large-scale teacher model (called ”soft target”) to impart knowledge to the lightweight student model. Due to the associations between different classes harboring in soft targets, the student model can learn the knowledge from the teacher model better than using one-hot labels [9]. Inspired by the related works in the above field, we propose a novel semi-supervised segmentation framework with Soft Pseudo-label Fusion, called ”SPFSeg”. The core of SPFSeg is establishing the effective soft pseudo-labels with the association knowledge of ROIs and backgrounds to narrow the image property gap between natural and medical images when conducting the pseudo-labeling paradigm, helping the segmentation model learn better object pattern understanding of ROIs and backgrounds. Our contributions are summarized as follows:
-
Inspired by ”soft target” in knowledge distillation, we design a new pseudo-labeling strategy called ”Soft Pseudo-label Fusion”. It integrates the ideas of ensemble learning and entropy minimization to generate the refine soft pseudo-labels, harboring the association knowledge of ROIs and backgrounds while maintaining the ”low-density” assumption.
-
Based on Soft Pseudo-label Fusion, we further develop a semi-supervised medical imaging segmentation framework called SPFSeg, to help the segmentation model learn a better pattern understanding in medical images. The teacher-student architecture with strong and weak augmentation effectively couples the proposed Soft Pseudo-label Fusion strategy, making SPFSeg exhibit remarkable performance on medical images with different modalities.
-
Extensive experiments have been conducted to evaluate SPFSeg on CXR-2014 [2], ISIC-2017 [10], and BUL-2020 [11] under different experimental settings. The results and qualitative analyses show that its performance can surpass that of existing SSL segmentation frameworks. In the case of using exceedingly limited labeled images, its segmentation performance outperforms other compared frameworks by a large margin.
The rest of this paper is organized as follows. We review the related works in knowledge distillation, pseudo-labeling, and entropy minimization in Sec. 2. The methodology of SPFSeg, including the overview, Soft Pseudo-label Fusion, and training objective, are presented in Sec. 3. The experiments and discussions are detailed in Sec. 4. Finally, our paper is concluded in Sec. 5.
2 Related works
2.1 Soft targets in knowledge distillation
Since large-scale models can excavate more information from the training dataset while lightweight models are more efficient and suitable for deployment, Knowledge distillation (KD) is proposed to transfer the abundant information learned from a large-scale model (the teacher model) to a lightweight model (the student model), achieving the goal of obtaining a lightweight model with comparable performance as a large-scale model [8]. One of the prevalent paradigms of KD is known as soft targets [9], which constrains the soft logits predicted by the last output layer of the student model to be as consistent as possible with those of the teacher model. This paradigm effectively exploits the informative dark knowledge contained in soft logits, which reveals the association between each class. Motivated by soft targets in KD, we introduce the soft targets into SSL of medical imaging segmentation, aiming at better representing the ambiguous regions commonly in medical images. Note that the teacher and student models in SPFSeg differ from those in KD. In our framework, the teacher model shares the same network architecture as the student, and it is momentum-updated by the weights of the student. It has been proved that the teacher model can output a robust supervision signal for the student model and make it perform well [12].
2.2 Pseudo-labeling in medical imaging segmentation
Although Deep Learning techniques have helped computer-aided medical imaging segmentation achieve significant strides, its data-hungry property still hinders its widespread applications in practical scenarios. Therefore, SSL has been gradually introduced into this field to make the segmentation model generalize well on unseen medical images [6, 13, 14]. Pseudo-labeling [4], derived from SSL of natural images, is a popular direction in SSL of medical segmentation. Specifically, it assigns fake pixel-level labels for each unlabeled image, and then combines them with annotated images to iteratively train the model. Many studies generated one-hot pseudo-labels using a fixed confidence-based threshold (similar to the operation in FixMatch [15]) or an adaptive threshold based on the learning ability or performance [16,17,18] to generate one-hot pseudo-labels. However, compared with natural images, many medical images’ pixels are difficult to be identified as a specific category in many cases. Using one-hot pseudo-labels is hard to represent them and thus limits the quality of the supervision for unlabeled images. In contrast to these frameworks, SPFSeg constructs soft pseudo-labels without threshold partition, aiming at guiding the segmentation model to learn the underlying association knowledge (i.e., ROIs and background). Moreover, we integrate the teacher-student architecture and differentiated perturbation (strong and weak augmentation) widely applied in consistency learning, encouraging the model to learn more essences of ROIs and backgrounds’ representations.
2.3 Entropy minimization in pseudo-labeling
In SSL, a popular assumption is that the classifier’s decision boundary should not pass-through high-density regions in the feature space, called the ”low-density” assumption [19]. Pseudo-labeling implicitly achieves this assumption via entropy minimization. It produces hard (one-hot) pseudo-labels from confident predictions and uses them as training targets via Cross Entropy loss, encouraging the model to output more low-entropy (confident) predictions on unlabeled data. In SPFSeg, we modify the vanilla pseudo-labeling to the soft pseudo-label. Although using soft labels can guide the model to learn underlying associations with ROIs and backgrounds, it may undermine the ”low-density” assumption and cause the degradation of segmentation performance. Thus, we couple the sharpening operation into Soft Pseudo-label Fusion to reconcile the target distribution for unlabeled data and reduce the entropy of soft pseudo-label. This operation has been proven to be essential in improving the performance of SPFSeg.
3 Methodology
3.1 Overview of SPFSeg
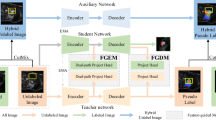
Figure 2 presents the overview of SPFSeg, which adopts multi-branch teacher-student architecture with Soft Pseudo-label Fusion strategy. The student model 𝜃 is updated by the supervision signal, and the teacher model \(\theta ^{\prime }\) is momentum updated by the weights of the student through Exponential Moving Average (EMA). The goal of SPFSeg is to train a semantic segmentation model by utilizing a tiny set of labeled images and a large number of unlabeled images. In every training step, a batch of N labeled images and corresponding labels \(\left \{{x_{l}^{n}}, {y_{l}^{n}}\right \}_{n=1}^{N}\), and N unlabeled images \(\{{x_{u}^{n}}\}_{n=1}^{N}\) are randomly sampled from the training dataset.
For N labeled images and their corresponding labels \(\left \{{x_{l}^{n}}, {y_{l}^{n}}\right \}_{n=1}^{N}\), they are first perturbed using strong augmentation strategy (detailed in Sec. 4.2): \(\tilde {x}_{l}^{n}={\Phi }\left ({x_{l}^{n}}\right )\) and \(\tilde {y}_{l}^{n}={\Phi }\left ({y_{l}^{n}}\right )\). Then, \(\{\tilde {x}_{l}^{n}\}_{n=1}^{N}\) are sent to the student model to get their predictions: \(Y\left (\tilde {x}_{l}^{n}\right )=f\left (\tilde {x}_{l}^{n}, \theta \right ), n \in [1,N]\). Finally, the labeled supervision loss is calculated between \(\tilde {y}_{l}^{n}\) and \(Y\left (\tilde {x}_{l}^{n}\right )\) using masked Cross Entropy, which will be formulated in Sec. 3.3.
For N unlabeled images \(\{{x_{u}^{n}}\}_{n=1}^{N}\), we use the proposed Soft Pseudo-label Fusion to generate their supervision, bridging the image property gap between natural and medical images when using Pseudo-labeling. Specifically, each image is randomly perturbed K times using strong and weak augmentation strategy (detailed in Sec. 4.2), respectively. In terms of each image \({x_{u}^{n}}\), K weakly perturbed versions are randomly generated using weak augmentation: \(\left (\hat {x}_{u}^{n}\right )^{i}={\Omega }\left ({x_{u}^{n}}\right ), i \in [1, K]\), and its K strongly perturbed versions are randomly generated using strong augmentation: \(\left (\tilde {x}_{u}^{n}\right )^{i}={\Phi }\left ({x_{u}^{n}}\right ), i \in [1, K]\). Then, the weakly perturbed versions are sent to the teacher model to get their predictions: \(\hat {Y}\left (\hat {x}_{u}^{n}\right )^{i}=f(\left (\hat {x}_{u}^{n}\right )^{i}, \theta ^{\prime })\), and the strongly perturbed versions are sent to the student model to get their predictions: \(\tilde {Y}\left (\tilde {x}_{u}^{n}\right )^{i}=f(\left (\tilde {x}_{u}^{n}\right )^{i}, \theta )\). Further, \(\hat {Y}\left (\hat {x}_{u}^{n}\right )^{i}\) are applied Soft Pseudo-label Fusion to generate the final soft pseudo-label \(\tilde {Y}\left (\hat {x}_{u}^{n}\right )^{i}\) for each \(\tilde {Y}\left (\tilde {x}_{u}^{n}\right )^{i}\). This strategy will be described in Sec. 3.2. Finally, the unlabeled supervision loss is calculated between \(\tilde {Y}\left (\hat {x}_{u}^{n}\right )^{i}\) and \(\tilde {Y}\left (\tilde {x}_{u}^{n}\right )^{i}\) using masked Mean Square Error, which will be formulated in Sec. 3.3.
In the training process of the student model, the teacher will also evolve to be more robust and generalized, serving as a better teacher for the student. When leveraging the unlabeled images, it will output diversified predictions to generate refined soft pseudo-labels through Soft Pseudo-label Fusion, which further provides potent supervision for unlabeled images.
3.2 Soft pseudo-label fusion
3.2.1 Inverse transformation
For the unlabeled images, Soft Pseudo-label Fusion utilizes the teacher’s multiple predictions under various perturbations to generate refined pseudo-labels for the student. Since these images are randomly perturbed by the weak augmentation strategy composed of various affine transformations (e.g., flipping, rotation, and scaling), their predictions are in different coordinate systems. Therefore, we first apply inverse transformations to eliminate the differences of weak augmentation, making the teacher predictions share the same coordinate system for subsequent soft pseudo-label fusion. In each process of weak augmentation, we calculate its inverse matrix to apply inverse transformations to its perturbed version’s prediction. For the prediction \(\hat {Y}\left (\hat {x}_{u}^{n}\right )^{i}, i \in [1, K]\), its inverse transformation version \(Y\left (\hat {x}_{u}^{n}\right )^{i}\) is calculated as follows:
where \(R^{-1}\left (\hat {x}_{u}^{n}\right )^{i}\) is the corresponding inverse transformation matrix applied in the i-th weak perturbation of \(\hat {x}_{u}^{n}\).
3.2.2 Fusion and refinement
As depicted in Fig. 3, for each unlabeled image, its \(Y\left (\hat {x}_{u}^{n}\right )^{i}, i \in [1, K]\) are first mapped into a categorial distribution \(P\left (\hat {x}_{u}^{n}\right )^{i}\) using channel-wise Softmax function, and then taken the average of them as follows:
Then, sharpening operation [20] is applied to reconcile \(\bar {P}\left (\hat {x}_{u}^{n}\right )\). Suppose pj is one of the pixel-wise softmax probability values of \(\bar {P}\left (\hat {x}_{u}^{n}\right )\) at the j-th channel, its sharpened probability value \(p_{j}^{\prime }\) is calculated as follows:
where C is the total number of channels (i.e., number of classes), and T is the sharpening temperature that controls the probability distribution of each class. Next, the sharpened \(\bar {P}\left (\hat {x}_{u}^{n}\right )\) is converted from categorial probabilities to the activation value \(\bar {Y}\left (\hat {x}_{u}^{n}\right )\). Suppose yj is the j-th converted activation value of \(p_{j}^{\prime }\), it is calculated as follows:
where eps is set to 1e− 7 to ensure the numerical stability. Finally, \(\bar {Y}\left (\hat {x}_{u}^{n}\right )\) is applied channel-wise normalization to rescale different values to a common scale. For the j-th converted activation value yj, its normalized value \(y_{j}^{\prime }\) is calculated as follows:
3.2.3 Soft pseudo-labeling
After getting each unlabeled image’s sharpened activation map \(\bar {Y}\left (\hat {x}_{u}^{n}\right )\), we use them to generate the soft pseudo-labels for the predictions of its strongly perturbed versions from the student model. We apply transformations to \(\bar {Y}\left (\hat {x}_{u}^{n}\right )\) based on respective transformation matrices applied in strong augmentations:
where \(R_{s}\left (\tilde {x}_{u}^{n}\right )^{i}\) is the transformation matrix of \(\tilde {x}_{u}^{n}\) in the i-th strong perturbation. The whole process of constructing the supervision for unlabeled images is shown in Algorithm 1.
3.3 Training objective
The training objective of SPFSeg is to minimize the total supervision loss, which is composed of two parts: the supervision loss of the labeled images Ls and the supervision loss of the unlabeled images Lu. Note that both strong and weak augmentations involve affine transformations, where some operations (e.g., scaling, rotation, and shearing) may generate undefined regions on images. In early experiments, we found that the segmentation model is sensitive to these undefined regions and tends to classify them as ROIs, which will impair the supervision of unlabeled images. Therefore, when calculating Ls and Lu, we set binary masks (the pixels in the valid region are marked as 1 and the pixels in the undefined region are marked as 0) to control where gradients should be passed through and thus neglect the supervision in the undefined region. The supervision loss of labeled images Ls is calculated by masked Cross Entropy, which is defined as follows:
where ℓce is the standard cross-entropy function and \(M\left (\tilde {x}_{l}^{n}\right )\) is the binary mask. If one value in \(M\left (\tilde {x}_{l}^{n}\right )\) is 0, its corresponding position in \(Y\left (\tilde {x}_{l}^{n}\right )\) will not be involved in the calculation of Ls. For the supervision of unlabeled images Lu, we adopt Mean Square Error (MSE) as it is a relatively stronger constraint compared to Cross Entropy loss [21]. The masked MSE is defined as follows:
where ℓmse is the standard MSE function, and \(M\left (\tilde {x}_{l}^{n}\right )^{i}\) is the binary mask for the i-th strongly perturbed version of \({x_{l}^{n}}\). Finally, the total supervision loss is calculated as follows:
where λ is a time-dependent weight used to rescale Lu. Since the student and teacher models are not reliable and robust at the beginning of model training, the supervision of Lu is of low quality. Hence, we set a warm-up stage to gradually increase λ based on Gaussian ramp-up function [12] at the early training stage:
where λmax is the maximum of λ, t is the number of current training steps, and tmax is the maximum of ramp-up length.
4 Experiments and discussions
4.1 Evaluation datasets
We evaluated the proposed framework on various medical image datasets, including Chest X-Ray of Tuberculosis dataset (CXR-2014) [2], International Skin Images Collaboration 2017 (ISIC-2017) [22], and Breast Ultrasound dataset (BUL-2020) [11]. The samples of the above datasets are shown in Fig. 4. The division protocols of these datasets are presented in Table 1.
1) CXR-2014. The Chest X-Ray of Tuberculosis dataset is provided by the National Library of Medicine, Maryland, USA, in collaboration with Shenzhen No.3 People’s Hospital, Guangdong Medical College, Shenzhen, China. This dataset contains raw 800 chest frontal x-rays images, where 704 images are annotated by Rajaraman et al. [23] and Computer Engineering Department, Igor Sikorsky Kyiv Polytechnic Institute, National Technical University of Ukraine. In this paper, we split the 704 annotated images to train and validate the segmentation performance.
2) ISIC-2017. The dataset of International Skin Images Collaboration - 2017 skin lesion challenge is to help participants develop image analysis tools that enable the automated diagnosis of melanoma from dermoscopic images. The lesion data includes the original image, paired with the expert manual tracing of the lesion boundaries in the form of a binary mask. This dataset contains 2000 training images, 150 validation images, and 600 testing images.
3) BUL-2020. The Breast Ultrasound Dataset (BUL) was created by Baheya Hospital for Early Detection & Treatment of Women’s Cancer, Cairo, Egypt, which is open-sourced in Kaggle Community (https://www.kaggle.com/datasets/aryashah2k/breast-ultrasound-images-dataset). These images are categorized into three classes, which are normal, benign, and malignant. It contains 780 images with an average image size of 500*500 pixels. In this dataset, multiple lesion annotations from the same subject are saved in independent files. Therefore, we merge all lesion annotations of each subject into one file in our experiment.
4.2 Implementation details
In all experiments, we adopt the Adam optimizer with step learning rate decay schedule (gamma = 0.8 for every 80 steps). The initial learning rate is 5e− 4, and the weight decay is 1e− 5 on ISIC-2017, 3e− 5 on CXR-2014 and BUL-2020. Every training batch consists of 24 annotated images and 24 unannotated images. All images are resized to 224*224. According to the segmentation difficulty of datasets, we select 1%, 3%, and 5% labeled data on CXR-2014, 1%, 5%, and 10% on ISIC-2017, and 5%, 10%, 20% on BUL-2020. All the experiments are trained with 800 iterations (except for 400 iterations when using 1% labeled data on CXR-2014 and ISIC-2017, and 5% on BUL-2020) and evaluated on one NVIDIA RTX 3090 GPU (24GB). The backbone segmentation network is adopted DeepLab v3+ [24] with ResNet-101 pretrained model.
For SPFSeg, the warm-up length tmax is set to 200 steps, and the maximum of \(\lambda ^{\prime }\) is set to 1.0. Sharpening temperature T is set to 0.5 on CXR-2014, 0.2 on BUL-2020, and 0.2 on ISIC-2017. In the perturbation process, strong and weak augmentation follows the strategy used in RandAugment [25]. The weak augmentation includes random flipping (not applied on CXR-2014), scaling, rotation (0∘, 90∘, 180∘, and 270∘), and shearing. Based on weak augmentation operations, strong augmentation additionally includes random color distortions implemented on brightness, contrast, saturation, hue, and Gaussian blur. The maximum color distortion degree is 1.0 on CXR-2014, 0.7 on ISIC-2017 and BUL-2020. The EMA decay is 0.97 on ISIC-2017, and 0.99 on CXR-2014 and BUL-2020. Note that the annotations of images are not applied random color distortions in strong augmentation.
4.3 Comparison with existing alternatives
We compared SPFSeg’s segmentation performance with other state-of-the-art semi-supervised frameworks (including Mean Teacher [12] (MT), FixMatch [15] (FM), and Cross Pseudo Supervision [26] (CPS)) in terms of Dice score and Jaccard Index. Considering the small number of datasets and the limited labeled images in experimental settings, the performance is prone to be unstable due to the sample distribution of the training set and labeled data. Therefore, all frameworks were trained in five rounds with the same random division protocol and took the average of metrics as the final results. The results are presented in Table 2, 3 and 4. A corresponding paired t-test (Table 5) was conducted to investigate the statistical significance between the results of SPFSeg and other compared SSL frameworks. The significance threshold was set to 0.05, where a p-value less than 0.05 indicates the statistical significance of the results. In our experiments, many cases were less than 0.01, implying that the results from SPFSeg were distinct from those of other frameworks.
Specifically, we can find that SPFSeg achieves the most significant performance improvement over the model trained with only limited labeled images, compared with other prior arts of SSL. On CXR-2014, all frameworks perform relatively well when using 3% and 5% labeled images because of simple object patterns (i.e., lungs). However, in the case of using 1% labeled images, SPFSeg has a 2.48% and 2.31% improvement in terms of Dice Score and Jaccard Index compared to the prior arts. Similarly, SPFSeg surpasses prior arts by a substantial margin, ranging from 1.12% to 2.35% in Dice Score and from 1.03% to 1.63% in Jaccard Index. Furthermore, on BUL-2020, the SPFSeg model continues to manifest its superiority, with an improvement of 1.09% to 2.16% in Dice Score and 0.57% to 1.43% in Jaccard Index when trained with 5% and 10% labeled images. Despite a slightly lower Dice Score compared to CPS when trained with 20% labeled images, SPFSeg still outperforms most of the prior arts in terms of overall performance.
Interestingly, compared to prior arts of SSL, SPFSeg has more significant performance gain when exceedingly limited labeled data are provided (i.e., 1% on CXR-2014 and ISIC-2017, and 5% on BUL-2020). We reckon that other frameworks’ outputs of the perturbed images are prone to have high variances when using exceedingly limited labeled data. This case will make the supervision of unlabeled images unstable, thus deteriorating the final segmentation performance. For SPFSeg, it generates multiple soft pseudo-labels and further integrates them to lower the variance of the strongly perturbed images’ pseudo-labels, which can act as a better supervision signal for unlabeled images. Moreover, we compare the performance of existing fully supervised segmentation methods. Table 6 shows that SPFSeg’s performance can even approach or outperform these methods with few labeled data on these datasets, showing an encouraging prospect for practical applications.
From the qualitative results shown in Fig. 5, we can observe that the ROIs segmented by SPFSeg are generally closer to their ground truths than those of the prior arts in the case of using 1% labeled data (except 5% on BUL-2020) as well as the same backbone network. On ISIC-2017, it maintains good segmentation performance under various cases, while other frameworks output some wrong regions due to the background noises (e.g., Row #1 and Row #2). In the challenging case of Row #4 where the ROI and background are visually similar, the proposed framework still outputs the most complete prediction, showing its robust segmentation performance in diverse subjects. On CXR-2014, the performances of prior arts are generally acceptable, except for some flaws in the upper and lower margins of the lungs. In contrast, SPFSeg segments the ROIs with greater precision and fineness on the margins. On BUL-2020, the presence of noise and artifacts in CT scans results in a relatively poor performance of all semi-supervised frameworks. Despite this, SPFSeg still performs better in terms of coverage area and the fineness of segmented regions.
Qualitative comparisons on CXR-2014, ISIC-2017, and BUL-2020 when trained with 1% labeled data (5% labeled data on BUL-2020). The “red” area is the ground truth that the framework does not predict, the “green” area is the wrong region predicted by the framework, and the “yellow” area is the overlapping area of the prediction result and the corresponding ground truth
4.4 Ablation studies
We conducted the ablation study to investigate the influence of Soft pseudo-label (SP), Soft pseudo-label fusion (SPF), teacher-student architecture (T&S), and strong and weak augmentation (SWA) in SPFSeg. The details of these sub-models with ablated parts are as follows: 1) the ablated variant of SP uses hard pseudo-labels to produce supervision for the unlabeled images; 2) the ablated variant of SPF is removed Eq. (2) \( \sim \) Eq. (6); 3) the teacher model of the ablated variant of T&S shares the weights of the student (gradients are only back-propagated on the student model); and 4) the ablated variant of SWA is only applied a simple augmentation strategy (only random flipping) to support SPF. The results of our ablation experiments are shown in Table 7. All results are obtained by training three rounds with 5% labeled data on ISIC-2017.
Soft Pseudo-label Fusion can better leverage the unlabeled images to build unlabeled supervision.
SPFSeg adopts the proposed Soft Pseudo-label fusion (SPF) to build the supervision of unlabeled images. It incorporates the association knowledge of ROIs and backgrounds into pseudo-labels and meanwhile guarantees the ”low-density” assumption. After replacing hard pseudo-labels in the ablated variant of SP, it has a 2.25% performance decline, demonstrating the effectiveness of imparting the underlying association knowledge to the segmentation model. Moreover, after removing complete SPF strategy, the ablated variant of SPF experiences a severe performance degradation of 3.62%. This indicates that SPF can generate better soft pseudo-labels for the segmentation model by fusing diversified predictions under different perturbations.
The momentum-updated teacher improves the quality of the supervision of unlabeled images.
In SPFSeg, we use EMA to momentum-update the teacher in each training step based on previous student models’ weights. After removing the teacher in SPFSeg, we can find the ablated version of T&S has a performance drop of 0.92%. This shows that momentum-updated teacher can output more robust predictions of weakly perturbed images. These images subsequently generate better soft pseudo-labels for the supervision of unlabeled images.
Strong and weak augmentation helps the model learn better representations of medical images.
In SPFSeg, strong and weak augmentation strategy is applied to the training images for the teacher and student. The removal of SWA results in a significant drop in performance of 3.32 %, highlighting its crucial role in helping SPFSeg learn better feature representations. On the one hand, the use of weak augmentation allows for the generation of diversified yet high-quality predictions from the teacher model (as shown in Fig. 6), which are necessary for producing better soft pseudo-labels. On the other, the supervision of unlabeled images will encourage the student model to output more consistent predictions with soft pseudo-labels under strong perturbations. Further, we also explore the performance influence of random augmentation operations (see Table 8). We can find that these operations are beneficial to improve the Dice score and Jaccard index of SPFSeg. Among them, color distortion is the most effective operation for CXR-2014 and ISIC-2017, and random scaling and shearing is the most effective operation for BUL-2020.
Visualization of the weakly perturbed images’ predictions on ISIC-2017. The predictions are undergone inverse transformations (see Sec. 3.2.1) to share the same coordinate space as the original image
4.5 Hyperparameter analysis
In SPFSeg, two hyperparameters are crucial to SPFSeg. One is the branch number K. In the teacher and student branches, we perturbed each image K times using strong and weak augmentation, respectively. A suitable setting of K guides the model to generate a certain number of predictions under various transformations, contributing to obtaining a high-quality soft pseudo-label through Soft Pseudo-label Fusion. However, an excessively large K is prone to bring more noise in Soft Pseudo-label Fusion, producing the adverse effects of pseudo-labels for unlabeled images. The other one is the sharpening temperature T, which reconciles the categorial distribution of the pseudo-labels. A larger T can impose a stronger constraint to maintain the ”low-density” assumption, while a smaller T can preserve more association knowledge. Therefore, in this section, we discuss to choose an appropriate branch number to get exquisite soft pseudo-labels and balance the low-entropy degree and the informativeness of the association knowledge.
As shown in Fig. 7, we evaluate the above two hyperparameters under different experimental settings on ISIC-2017 (10% labeled data). The results indicate that SPFSeg achieves the best segmentation performance when K = 2. With the increase of K, its performance experiences a degradation. We reckon that overmuch predictions of the teacher model bring too many noises, which is not contributing to producing ideal soft pseudo-labels for the student. In terms of the sharpening temperature T, SPFSeg yields the best segmentation performance when T = 0.2. It is a relatively low temperature, which can make pseudo-labels to be very low-entropy. This suggests that the premise of introducing association knowledge of ROIs and backgrounds is to ensure that the predictions are low-entropy. Introducing too much association knowledge while neglecting the ”low-density” assumption will cause the degradation of segmentation performance.
5 Conclusion
In this paper, we propose a novel semi-supervised medical imaging segmentation framework through soft pseudo-label fusion, SPFSeg. Its core idea is utilizing the dark knowledge, i.e., the association knowledge between ROIs and backgrounds, to bridge the image property gap between natural and medical images, making pseudo-labeling fit for medical imaging segmentation better. SPFSeg is integrated strong and weak augmentation with the teacher-student architecture to output multiple predictions under various transformations simultaneously. These outputs from the teacher are then integrated to generate refined soft pseudo-labels with low entropy for the student, helping SPFSeg establish potent supervision for unlabeled images. Extensive experiments show significant improvement in semi-supervised medical imaging segmentation performance on CXR-2014, ISIC-2017, and BUL-2020. One demerit of SPFSeg is that it has high computation and memory costs due to multiple inferences in the Soft Label Fusion stage. In the future, we would like to investigate possible alternatives to overcome the above problem and transfer this framework to 3D medical imaging to validate its applicability.
References
Berseth M. (2017) ISIC 2017 - skin lesion analysis towards melanoma detection. CoRR. abs/1703.00523
Jaeger S., Candemir S., Antani S., Wáng Y.-X.J., Lu P.-X., Thoma G. (2014) Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quantitative imaging in medicine and surgery 4(6):475
Bernard O., Lalande A., Zotti C., Cervenansky F., Yang X., Heng P.-A., Cetin I., Lekadir K., Camara O., Ballester M.A.G., et al. (2018) Deep learning techniques for automatic mri cardiac multi-structures segmentation and diagnosis: is the problem solved?. IEEE transactions on medical imaging 37(11):2514–2525
Lee D.-H. (2013) Pseudo-label: The simple and efficient semi-supervised learning method for deep neural networks. In: Workshop on Challenges in Representation Learning, ICML, vol. 3, p. 2
Kamraoui R.A., Ta V.-T., Papadakis N., Compaire F., Manjon J.V., Coupé P. (2021) Popcorn: Progressive pseudo-labeling with consistency regularization and neighboring. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 373–382. Springer
Li Y., Chen J., Xie X., Ma K., Zheng Y. (2020) Self-loop uncertainty: A novel pseudo-label for semi-supervised medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 614–623. Springer
Liu F., Tian Y., Chen Y., Liu Y., Belagiannis V., Carneiro G. (2022) Acpl: Anti-curriculum pseudo-labelling for semi-supervised medical image classification. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 20697–20706
Gou J., Yu B., Maybank S.J., Tao D. (2021) Knowledge distillation: A survey. Int J Comput Vision 129(6):1789–1819
Hinton G., Dean J., Vinyals O. (2014) Distilling the knowledge in a neural network. In: NIPS
Li Y., Shen L. (2018) Skin lesion analysis towards melanoma detection using deep learning network. Sensors 18(2):556
Al-Dhabyani W., Gomaa M., Khaled H., Fahmy A. (2020) Dataset of breast ultrasound images. Data in brief 28:104863
Laine S., Aila T. (2017) Temporal ensembling for semi-supervised learning.. In: ICLR (Poster)
Luo X., Chen J., Song T., Wang G. (2021) Semi-supervised medical image segmentation through dual-task consistency. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 35, pp. 8801–8809
You C., Zhou Y., Zhao R., Staib L., Duncan J.S. (2022) Simcvd: Simple contrastive voxel-wise representation distillation for semi-supervised medical image segmentation. IEEE Transactions on Medical Imaging
Sohn K., Berthelot D., Carlini N., Zhang Z., Zhang H., Raffel C.A., Cubuk E.D., Kurakin A., Li C.-L. (2020) Fixmatch: Simplifying semi-supervised learning with consistency and confidence. Adv Neural Inf Process Syst 33:596–608
Zhang W., Zhu L., Hallinan J., Zhang S., Makmur A., Cai Q., Ooi B.C. (2022) Boostmis: Boosting medical image semi-supervised learning with adaptive pseudo labeling and informative active annotation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 20666–20676
Lai Z., Wang C., Hu Z., Dugger B.N., Cheung S.-C., Chuah C.-N. (2021) A semi-supervised learning for segmentation of gigapixel histopathology images from brain tissues. In: 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), pp. 1920–1923. IEEE
Thompson B.H., Di Caterina G., Voisey J.P. (2022) Pseudo-label refinement using superpixels for semi-supervised brain tumour segmentation. In: 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), pp. 1–5. IEEE
Chapelle O., Zien A. (2005) Semi-supervised classification by low density separation. In: International Workshop on Artificial Intelligence and Statistics, pp. 57–64. PMLR
Berthelot D., Carlini N., Goodfellow I., Papernot N., Oliver A., Raffel C. (2019) Mixmatch: A holistic approach to semi-supervised learning. In: NeurIPS
Brier G.W. (1950) Verification of forecasts expressed in terms of probability. Monthly weather review 78(1):1–3
Codella N.C., Gutman D., Celebi M.E., Helba B., Marchetti M.A., Dusza S.W., Kalloo A., Liopyris K., Mishra N., Kittler H., et al. (2018) Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 168–172. IEEE
Rajaraman S., Folio L.R., Dimperio J., Alderson P.O., Antani S.K. (2021) Improved semantic segmentation of tuberculosis—consistent findings in chest x-rays using augmented training of modality-specific u-net models with weak localizations. Diagnostics 11(4):616
Chen L.-C., Zhu Y., Papandreou G., Schroff F., Adam H. (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the European Conference on Computer Vision (ECCV), pp. 801–818
Cubuk E.D., Zoph B., Shlens J., Le Q.V. (2020) Randaugment: Practical automated data augmentation with a reduced search space. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, pp. 702–703
Chen X., Yuan Y., Zeng G., Wang J. (2021) Semi-supervised semantic segmentation with cross pseudo supervision. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 2613–2622
Zhang Z., Fu H., Dai H., Shen J., Pang Y., Shao L. (2019) Et-net: A generic edge-attention guidance network for medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 442–450. Springer
Qin X., Xu M., Zheng C., He C., Zhang X. (2021) Multi-scale feedback feature refinement u-net for medical image segmentation. In: 2021 IEEE International Conference on Multimedia and Expo (ICME), pp. 1–6. IEEE
Milletari F., Rieke N., Baust M., Esposito M., Navab N. (2018) Cfcm: segmentation via coarse to fine context memory. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 667–674. Springer
Peng T., Gu Y., Ye Z., Cheng X., Wang J. (2022) A-lugseg: Automatic and explainability-guided multi-site lung detection in chest x-ray images. Expert Systems with Applications, 116873
Schlemper J., Oktay O., Schaap M., Heinrich M., Kainz B., Glocker B., Rueckert D. (2019) Attention gated networks: Learning to leverage salient regions in medical images. Medical image analysis 53:197–207
Lei B., Xia Z., Jiang F., Jiang X., Ge Z., Xu Y., Qin J., Chen S., Wang T., Wang S. (2020) Skin lesion segmentation via generative adversarial networks with dual discriminators. Med Image Anal 64:101716
Wu H., Chen S., Chen G., Wang W., Lei B., Wen Z. (2022) Fat-net: Feature adaptive transformers for automated skin lesion segmentation. Med Image Anal 76:102327
Wang Y., Deng Z., Hu X., Zhu L., Yang X., Xu X., Heng P.-A., Ni D. (2018) Deep attentional features for prostate segmentation in ultrasound. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 523–530. Springer
Xue C., Zhu L., Fu H., Hu X., Li X., Zhang H., Heng P.-A. (2021) Global guidance network for breast lesion segmentation in ultrasound images. In: Medical image analysis, vol 70, p 101989
Wang K.-N., Yang X., Miao J., Li L., Yao J., Zhou P., Xue W., Zhou G.-Q., Zhuang X., Ni D. (2022) Awsnet: An auto-weighted supervision attention network for myocardial scar and edema segmentation in multi-sequence cardiac magnetic resonance images. Medical Image Analysis, 102362
Acknowledgments
This work is supported in part by Shanghai science and technology committee under grant No. 21511100600. We appreciate the support of the High Performance Computing Center of Shanghai University, and Shanghai Engineering Research Center of Intelligent Computing System.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, X., Wu, Y. & Dai, S. Semi-supervised medical imaging segmentation with soft pseudo-label fusion. Appl Intell 53, 20753–20765 (2023). https://doi.org/10.1007/s10489-023-04569-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-023-04569-6