Abstract
The Gram-staining negative, oxidase and catalase negative strain KC-ST17T, isolated from saline-alkali land, was characterized using a polyphasic approach to determine its taxonomic position. Using 16S rRNA gene sequence analysis, the highest similarity of strain KC-ST17T was found with Nitratireductor pacificus CCTCC AB 209302T (97.2%). Cells are aerobic, non-motile, and rod-shaped. The isolate was found to be able to grow in NaCl concentrations of 0–4.0%. The assembled genome of strain KC-ST17T had a total length of 4.9 Mb with a G + C content of 62.7%. According to genome analysis, strain KC-ST17T encodes genes involved in the reduction of nitrate to nitrite, which may play a role in the utilization of nitrogenous compounds from the soil as an immediate source of energy. Based on the phenotypic characteristics and phylogenetic analysis, strain KC-ST17T was confirmed to represent a novel species in the Nitratireductor genus; thus, the name Nitratireductor luteus sp. nov. was proposed. The type strain of this species was KC-ST17T (= KCTC 92119T = MCCC 1K07309T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Nitratireductor was proposed by Labbé et al. (2004) and belongs to the family Phyllobacteriaceae, order Rhizobiales, class Alphaproteobacteria, and phylum ‘Pseudomonadota’. At the time of writing, the genus included ten validly published species with the correct name (https://lpsn.dsmz.de/genus/nitratireductor May 2022), and the type species was Nitratireductor aquibiodomus (Labbé et al. 2004). The type strains of this species have been isolated from various environmental habitats, including marine denitrification systems (Labbé et al. 2004), seaweed (Kang et al. 2009), black beach sand (Kim et al. 2009), deep-sea water (Lai et al. 2011), pyrene-degrading consortia (Lai et al. 2011), salt lakes (Yu et al. 2016) and estuaries (Ou et al. 2017), and diatom cultures (Jang et al. 2011). The bacteria of this genus are gram-negative, aerobic, rod-shaped or coccoid, with variable motility and nitrate-reducing abilities. The major quinone is Q-10 and the genomic DNA G + C content is 56.7–63.0%.
In this study, we analyzed the characteristics of a novel species using a series of cultivation techniques and genetic manipulations (Ramasamy et al. 2014) including phenotype identification, housekeeping gene sequencing, phylogenetic analysis, genome sequencing and annotation, fatty acid methyl ester analysis, and antibiotic susceptibility test. We propose to establish this strain as a representative of a novel species of the genus Nitratireductor, with the name Nitratireductor luteus sp. nov.
Materials and methods
Isolation and culture conditions
Strain KC-ST17T was originally isolated from saline-alkali land collected from the Akesu region (41°9′90″N, 80°30′80″E) in Xinjiang, China, in May 2021. Soil samples were suspended in sterile water and serially diluted. A 1:1000 dilution of the sample was cultivated on nutrient agar (NA) plates. The plates were incubated at 30 °C and checked for growth after 1–2 d. A single colony was picked and sub-cultured until it was pure. The pure strain was cultivated routinely on NA and NB media at 33 °C under aerobic conditions and then preserved at −80 °C in sterile 1.0% (w/v) saline supplemented with 15.0% (v/v) glycerol. Nitratireductor pacificus CCTCC AB209302T was obtained from the China Center for Type Culture Collection (CCTCC). The reference strain was cultured under the same conditions as strain KC-ST17T for comparative study.
Molecular analysis
Genomic DNA was isolated from a five-day-old NB liquid culture using a bacterial genomic DNA kit (Takara Bio, Shiga, Japan), following the manufacturer’s instructions. The 16S rRNA gene was amplified using PCR with the forward primer 27F and reverse primer 1492R, as described previously (Liu et al. 2014). Purified PCR products were sequenced by BGI Co. Ltd (Qingdao, China) and resulted in a 1309 bp almost complete 16S rRNA gene sequence. The 16S rRNA gene sequence was compared with sequences from the NCBI database using BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST) (Altschul et al. 1990), as well as with sequences available in the EZTaxon database (www.ezbiocloud.net/) (Yoon et al. 2017). We performed phylogenetic analysis based on the available sequences. A phylogenetic tree was constructed using the neighbor-joining algorithm implemented in the software package MEGA (version 7.0) (Kumar et al. 2016). Phylogenetic trees were also generated using the maximum-likelihood and maximum-parsimony algorithms. Evolutionary distances were calculated using Kimura’s 2-parameter method (Kimura et al. 1980), where gaps were completely deleted. Bootstrap analysis was performed with 1000 replications (bootstrap analysis; Felsenstein et al. 1985) to provide confidence estimates for tree topologies. Moreover, phylogenetic relationships based on nucleotide sequences were analyzed via UBCG (Na et al. 2018), and phylogenetic trees were constructed using FastTree (Price et al. 2010) with GTR + CAT parameters and IQTree (Trifinopoulos et al. 2016) with the GTR + F + I + G4 model and 1000 bootstrap replicates on the basis of 20 genomes.
Phenotypic characterization
To determine the morphological characteristics of the isolated strain, strain KC-ST17T was cultured in modified NA medium at 33 °C for 48 h. The shape, size, gloss, edge, and color of the colonies on the plate were recorded. Light microscopy (E600; Nikon USA, Melville, NY, USA) and transmission electron microscopy (TEM; Jem-1200; JEOL) were used to observe the cell phenotypes. Motility was determined using the hanging-drop method and gliding motility was determined as described by Bowman (Bowman et al. 2000). Gram staining was performed as previously described by Smibert and Krieg (1994). Growth at different temperatures (10, 15, 20, 25, 30, 33, 35, 37, 40, 42, 45, and 50 °C) and pH (5.5–9.0, at increments of 0.5 pH units) was determined in NB. Growth in media with different NaCl concentrations was investigated on basal medium with various NaCl concentrations (0.5, 1.5, 2.0, 2.5, 3.0, 3.5, 4.5, and 6.5%, w/v) (Pridham and Gottlieb 1948). The growth range and optimal pH and NaCl concentrations were determined by measuring the optical density (OD) at 600 nm (Krist et al. 1998).
Oxidase activity was tested using an oxidase reagent kit (BioMérieux, Marcy l’Etoile, France), according to the manufacturer’s instructions. Catalase activity was detected through bubble production using 3.0% (v/v) H2O2. The reduction of nitrate and hydrolysis of starch, casein, CM-cellulose, and Tween 40 and 80 were determined according to the methods described by Dong and Cai (Kanehisa et al. 2001). Growth under anaerobic (15% CO2 and 85% N2) condition was determined after incubation for 14 d in an anaerobic jar. Antibiotic susceptibility was investigated by the disc diffusion plate method (Bauer et al. 1966) using antibiotic discs on NA incubated for 7 d at 33 °C. Fourteen antibiotic discs were used (µg/disc, unless otherwise indicated): chloramphenicol (30), carbenicillin (100), penicillin (10), ceftriaxone (30), clarithromycin (15), erythromycin (15), tetracycline (30), gentamycin (10), tobramycin (10), vancomycin (30), streptomycin (10), lincomycin (2), streptomycin (10), and neomycin (30). Acid production from different carbon sources, assimilation of different substrates, and enzymatic activities of strain KC-ST17T were investigated using API 50 CH, API 20 NE, and API ZYM kits (BioMérrieux) according to the manufacturer’s instructions. The API 50 CH and 20 NE tests were performed after 24–48 h of incubation at 33 °C.
Chemotaxonomy
For cellular fatty acid analysis, strain KC-ST17T was grown on NA plates at 33 °C for 48 h (at the late exponential stage of growth). Cellular fatty acid methyl esters (FAMEs) were obtained from cells by saponification, methylation, and extraction, following the MIDI protocol. Cellular FAMEs were separated using gas chromatography (GC) (6890) and identified and quantified using the Sherlock Microbial Identification System (MIDI-6890 with the database TSBA6). The isoprenoid quinone of strain KC-ST17T was extracted from freeze-dried cell material using the two-stage method described by Tindall et al. (2007) and subsequently analyzed using HPLC (Hiraishi et al. 1996). Polar lipids were extracted using a chloroform/methanol system and analyzed using two-dimensional thin-layer chromatography, as described previously (Fang et al. 2017).
Genome sequencing, annotation, and analysis
For genome sequencing, the DNA was prepared using a bacterial DNA isolation kit (Takara). The draft genome of strain KC-ST17T was sequenced by MAGIGENE Biological Technology Co. Ltd. (Guangzhou, China) and assembled using hybrid assembly methods. Sequencing was carried out on an Illumina Hiseq Xten platform (Illumina Inc., San Diego, CA, USA) at Guangdong Magigene Biotechnology Co. LTD (Guangzhou, China) using the pair-end 150-bp sequencing protocol. Long reads were generated by SMRT sequencing using the Pacific Biosciences RS II sequencer (PacBio, Menlo Park, CA, USA) according to standard protocols. Low-quality reads were filtered using Mecat2 (https://github.com/xiaochuanle/MECAT2; Xiao et al. 2017) after sequencing and assembled using SMRT Link v5.1.0. The hybrid assembled genome was generated using Unicycler (https://github.com/rrwick/Unicycler; Wick et al., 2017). The assembled genome was polished by applying the arrow algorithm of the Genomic Consensus package (https://github.com/PacificBiosciences/GenomicConsensus).
Protein-encoding regions were identified using the Rapid Annotations in the Subsystem Technology (RAST) server (Aziz et al. 2008) and the Cluster of Orthologous Group of Proteins (COG) (Tatusov et al. 2003), and the genes were annotated using Koala (KEGG) (Kanehisa et al. 2016). Furthermore, the NCBI prokaryotic genome annotation pipeline server was used to identify the genes of strain KC-ST17T (Angiuoli et al. 2008). The average nucleotide identity (ANI) (RodríguezR and Konstantinidis 2016) and in silico digital DNA:DNA hybridization (DDH) were calculated using JSpecies WS (http://jspecies.ribohost.com/jspeciesws/) and the GGDC method, with the recommended formula 2, available at the TYGS web service (Meier-Kolthof and Göker 2019). Gene clusters potentially involved in the production of secondary metabolites were determined using antiSMASH 4.0 (Blin et al. 2017).
Results and discussion
Phylogenic analysis
According to comparisons with the 16S rRNA full-length gene sequences (1501 bp) in the EzTaxon database, the highest levels of sequence similarity occurred with N. pacificus CCTCC AB209302T (97.2%), N. aquibiodomus JCM 21793T (96.9%), and N. soli ZZ-1T (96.3%). The 16S rRNA gene-based phylogenetic trees suggest that strains SC-ST17T and N. pacificus CCTCC AB209302T form a distinct phyletic lineage within the Nitratireductor genus, and their relationship is shown in the neighbor-joining (NJ) phylogenetic tree (Fig. 1). Moreover, the overall topologies of the phylogenetic trees obtained with maximum likelihood (ML) and maximum parsimony (MP) were similar; therefore, it can be concluded that strain SC-ST17T belongs to the genus Nitratireductor. Moreover, the phylogenetic analyses based on a more comprehensive data set of validly published name strains genomes is presented in Fig. 2, and similar conclusion was also obtained. Therefore, the low sequence similarity and phylogenetic position on the trees indicated that strain SC-ST17T may represent a novel species in the genus Nitratireductor.
Neighbour-joining phylogenetic tree based on full-length 16S rRNA gene sequence (1501 bp), showing the phylogenetic position of strain KC-ST17T among members of the genus Nitratireductor. Numbers on nodes represent bootstrap values (NJ) based on 1000 replications. Only bootstrap values higher than 70.0% are marked on the branches. Filled circles indicate nodes also obtained in both maximum-likelihood and maximum-parsimony trees. Bar, 0.01 substitutions per nucleotide position
The phylogenetic trees based on based on nucleotide sequences from 20 genomes of related strains showed the taxonomic position of strain KC-ST17T. Filled circles indicate nodes overlapping on trees reconstructed using FastTree and IQTree algorithms. Numbers on nodes represent bootstrap values (FastTree/IQTree) based on 1000 replications. Bootstrap values (> 70.0%) based on 1000 replicates are shown at branch nodes. Bar, 0.005 substitutions per nucleotide position
Phenotypic and biochemical characterization
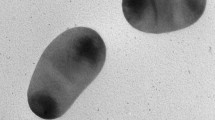
The basic features of strain SC-ST17T, including cell morphology, growth, and mechanism of cell division, are summarized in Table 1 and compared to those of N. pacificus CCTCC AB209302T, N. aquibiodomus JCM 21793T, and N. soli ZZ-1T. The morphological features of SC-ST17T cells harvested during the exponential growth phase were analyzed using TEM (Jem-1200; JEOL). Strain SC-ST17T forms rod-shaped cells of approximately 0.3–0.4 μm width and 0.6–1.2 μm length (Fig. S1). The cell size and shape of strain SC-ST17T are comparable to those of the known type species in the genus Nitratireductor. Cells of strain SC-ST17T were Gram-staining negative, oxidase and catalase negative as typical for members of the genus Nitratireductor. Casein, starch, CM-cellulose, Tween 40 and 80 are negative.
Cells are non-motile, grow at temperatures of 20–40 ℃, at a pH range of 6.5–9.0 and in 0–4.0% (w/v) NaCl. Optimal growth was observed at 30–33 ℃, 1.0% (w/v) NaCl and pH 7.0. Strain SC-ST17T and N. pacificus CCTCC AB209302T favour higher temperatures (40–42 ℃) than N. aquibiodomus JCM 21,793T (37 °C) or N. soli ZZ-1T (37 °C) (Table 1). With regard to pH range, strains SC-ST17T, N. pacificus CCTCC AB209302T, and N. soli ZZ-1T are able to tolerate slightly alkaline growth conditions (pH 9.0–10.0), whereas N. aquibiodomus JCM 21,793T require more neutral environments (pH 8.0). However, they all grow optimally under neutral conditions (pH 7.0–7.5). The NaCl concentration optimum of strain SC-ST17T (1.0%) is approaching N. soli ZZ-1T (1.0%); however, they all are slightly lower than N. pacificus CCTCC AB209302T (3.0%) and N. aquibiodomus JCM 21793T (2.5%). These differences likely reflect the different natural habitats in which the strains were isolated. N. pacificus CCTCC AB209302T was isolated from enriched sediment from the Pacific Ocean, whereas N. aquibiodomus JCM 21793T was isolated from the marine denitrification system, explaining the preference for NaCl concentrations. In contrast, strains SC-ST17T and N. soli ZZ-1T were isolated from soil. Meanwhile, it is also an important point to distinguish strain SC-ST17T from the closest neighbour N. pacificus CCTCC AB209302T.
Chemotaxonomic characteristics
The predominant cellular fatty acids in strain SC-ST17T are summed feature 8 (C18:1ω7c and/or C18:1ω6c, 55.3%) and C19:0 ω8c cyclo (17.3%). The fatty acid profile is similar to that of N. pacificus CCTCC AB209302T, which is in accordance with the description of the Nitratireductor genus. However, the ratios of the different components are different. The complete fatty acid compositions are shown in Table S3. The only respiratory quinone in the strain SC-ST17T is Q-10. Strain SC-ST17T exhibits a complex polar lipid profile consisting of diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), one phospholipid (PL), and one uncharacterized lipids (L) as the dominant elements. In comparison, N. pacificus CCTCC AB209302T differed from strain SC-ST17T by different contents of one phospholipid (PL) and an unidentified lipid (L2) (Fig. S2). Overall, it could be distinguished from on the basis of the chemotaxonomic characteristics of the new specie that distinguished it from the closely related species in the genus Nitratireductor.
Genome analysis
The complete genome size of strain SC-ST17T was 4,886,370 bp with DNA G + C content of 62.7%, which was consistent with the G + C content of the genus Nitratireductor. Similar genome features were observed for the closest neighbor N. pacificus CCTCC AB209302T (genome size 4,611,341 bp and G + C content 62.1%). The genome of strain SC-ST17T has a larger size than majority of the existing-sequenced strains, indicating substantial strain-to-strain variation. Annotation of the genome of strain SC-ST17T consisted of 4867 predicted protein-coding genes and 48 tRNAs, by contrast, N. pacificus CCTCC AB209302T possessed 4297 predicted protein-coding genes and 48 tRNAs. The genomic properties of strain SC-ST17T and other type strains within the genus Nitratireductor are summarized in Table S1. The average nucleotide identity (ANI) and in silico digital DDH (dDDH) values between strain SC-ST17T and strain N. pacificus MCCC 1A01024T were 76.2% and 33.8% (using GBDP distance formula 2), respectively. The ANI and dDDH values between strain SC-ST17T and other related species of the genus Nitratireductor were below the recommended thresholds of 95–96% and 70.0%, species demarcation (Ciufo et al. 2018; Meier-Kolthof and Göker 2019) (Table S2).
Based on the genomes of strain SC-ST17T and closely related species, we analyzed the number of putative carbohydrate-active enzymes and gene clusters putatively involved in the synthesis of secondary metabolites. These numbers can provide a first impression of the metabolic capabilities of the strain, for example, in competitive environments, in which complex polysaccharides (e.g., derived from macroscopic phototrophs) function as a major source of carbon and energy. The observed number of 124 putative carbohydrate-active enzymes of strain SC-ST17T is in the middle range to higher than that of its relatives, which harbor between 87 and more than 150 such enzymes. Strain SC-ST17T has the second largest genome of the four compared strains and is the strain with the second-lowest number of carbohydrate-active enzymes.
Although the difference in the genome size of strains SC-ST17T and N. pacificus CCTCC AB209302T was only approximately 0.28 Mb, the number of carbohydrate-active enzymes was nearly 1.2-fold different in a direct comparison of these two species. The number of proteins belonging to such classes is more likely a reflection of the complexity of the isolated environment, and perhaps, there is a certain correlation with the size of the genome. Meanwhile, the number of gene clusters putatively involved in the production of secondary metabolites was clearly correlated with genome size. Strain N. soli ZZ-1T had both the largest genome and the highest number of predicted clusters among the four strains.
Strain SC-ST17T can convert nitrate to nitrite but cannot reduce nitrite to nitrogen. The genome of strain SC-ST17T is also predicted to have genes involved in the reduction of nitrate to nitrite, potentially enabling strain SC-ST17T to utilize nitrogenous compounds from the soil as an immediate source of energy. The genome sequence of strain SC-ST17T and its curated annotation are important assets for better understanding its interaction with crops and other organisms in the soil environment, and will open up new opportunities in the functional analysis of this species in the global biogeochemical nitrogen cycle (Gu et al. 2013).
Conclusion
Taken together, strain SC-ST17T could be distinguished from the closely related type strains by several phenotypic characteristics (morphological and chemotaxonomic markers), especially with regard to optimum temperature, genome size, and DNA G + C content. The 16S rRNA gene sequence similarities to the closely related taxa and overall genome-related indices (ANI and dDDH) also indicate its distance from other species. These differences support the results of the phylogenetic inference and justify the delineation of strain SC-ST17T from previously described species in the genus Nitratireductor. Thus, we propose assigning the strain to a novel species of a novel genus, for which the name Nitratireductor luteus sp. nov. is proposed. The GenBank accession numbers of the 16S rRNA gene and genome sequences of strain SC-ST17T are OK143313 and JAKJPO000000000, respectively.
Description of Nitratireductor Luteus sp. nov.
Cells are gram-negative, aerobic, non-motile, rod-shaped, 0.3–0.4 μm in width, and 0.6–1.2 μm in length. Colonies on NA plates are circular, convex, smooth, pale-pigmented, and approximately 0.6–1.0 mm in diameter after 48 h at 33 °C. Growth occurs at 20–40 °C (optimum 30–33 °C), at pH 6.5–9.0 (optimum pH 7.0), and in the presence of 0–4.0% (w/v) NaCl (optimum 1.0%). The strain reduces nitrate to nitrite but does not produce nitrogen. It is catalase and oxidase-negative, negative for H2S production, indole production, gelatinase, Voges–Proskauer reaction, ONPG test, and Simmons citrate utilization. It does not hydrolyse starch, casein, CM-cellulose, Tween 40, and Tween 80. Acid is produced from d-ribose, l-sorbose, esculin ferric citrate, d-turanose, and d-tagatose, but not from glycogen, glucose, d-fucose, l-arbaitol, potassium 5-ketogluconate, d-raffinose, and d-ribose. The cells were positive for alkaline phosphatase, esterase (C4), esterase lipase (C8, weakly), leucine arylamidase, valine arylamidase (weakly), α-glucosidase (weakly), and N-acetyl-β-glucosaminidase (weakly). The tests for lipase (C14), trypsin, cystine arylamidase, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, α-mannosidase, and α-fucosidase were negative. The API 20 NE utilizes adipic acid (weakly), maltose, d-glucose, d-mannose (weakly), n-acetylglucosamine, and trisodium citrate, but not maltose, d-mannitol, l-arabinose, malic acid, phenylacetic acid, or potassium gluconate. Q-10 is the sole respiratory quinone. The major cellular fatty acids are summed feature 8 (C18:1ω7c and/or C18:1ω6c) and C19:0 ω8c cyclo. The major polar lipids are diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), one phospholipid (PL), and one uncharacterized lipid (L) as the dominant elements.
The type strain SC-ST17T (= KCTC 92,119T = MCCC 1K07309T) was isolated from saline-alkali land. The G + C content of the genomic DNA of strain SC-ST17T is 62.7%.
Data availability
The genome and 16S rRNA gene sequence are available from GenBank under the accession numbers provided in the manuscript.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Angiuoli SV, Gussman A, Klimke W, Cochrane G, Field D (2008) Toward an online repository of standard operating procedures (SOPs) for (Meta) genomic annotation. OMICS 12:137–141
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 9:75
Bauer A, Kirby W, Sherris J, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496
Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG (2017) AntiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45:W36–W41
Bowman JP (2000) Description of Cellulophaga algicola sp. nov., isolated from the surfaces of Antarctic algae, and reclassification of Cytophaga uliginosa (ZoBell and Upham 1944) reichenbach 1989 as Cellulophaga uliginosa comb. nov. Int J Syst Evol Microbiol 50:1861–1868
Chen Q, Ni HY, Zhuang W et al (2015) Nitratireductor soli sp. nov., isolated from phenol-contaminated soil. Antonie Van Leeuwenhoek 108:1139–1146
Ciufo S, Kannan S, Sharma S et al (2018) Using average nucleotide identity to improve taxonomic assignments in prokaryotic Genomes at the NCBI. Int J Syst Evol Microbiol 68:2386–2392
Dong X, Cai M (2001) Determination of biochemical characteristics. In:Dong XZ and Cai MY (editors). Manual for the Systematic Identification of General Bacteria. Beijing: Science Press; 2001. pp. 370–398
Fang DB, Han JR, Liu Y, Du ZJ (2017) Seonamhaeicola marinus sp. nov., isolated from marine algae. Int J Syst Evol Microbiol 67:4857–4861
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gu B, Chang J, Min Y et al (2013) The role of industrial nitrogen in the global nitrogen biogeochemical cycle. Sci Rep 3:2579
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by highperformance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Jang GI, Hwang CY, Cho BC (2011) Nitratireductor aquimarinus sp. nov., isolated from a culture of the diatom Skeletonema costatum, and emended description of the genus Nitratireductor. Int J Syst Evol Microbiol 61:2676–2681
Kanehisa M, Sato Y, Morishima K (2016) BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731
Kang HS, Yang HL, Lee SD (2009) Nitratireductor kimnyeongensis sp. nov., isolated from seaweed. Int J Syst Evol Microbiol 59:1036–1039
Kim KH, Roh SW, Chang HW, Nam YD, Yoon JH et al (2009) Nitratireductor basaltis sp. nov., isolated from black beach sand. Int J Syst Evol Microbiol 59:135–138
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Krist KA, Ross T, McMeekin TA (1998) Final optical density and growth rate; effects of temperature and NaCl differ from acidity. Int J Food Microbiol 34:195–203
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Labbe N, Parent S, Villemur R (2004) Nitratireductor aquibiodomus gen. nov., sp. nov., a novel alpha-proteobacterium from the marine denitrification system of the Montreal Biodome (Canada). Int J Syst Evol Microbiol 54:269–273
Lai Q, Yu Z, Yuan J, Sun F, Shao Z (2011) Nitratireductor indicus sp. nov., isolated from deep-sea water. Int J Syst Evol Microbiol 61:295–298
Lai Q, Yu Z, Wang J, Zhong H, Sun F et al (2011) Nitratireductor pacificus sp. nov., isolated from a pyrene-degrading consortium. Int J Syst Evol Microbiol 61:1386–1391
Liu QQ, Li XL, Rooney AP, Du ZJ, Chen GJ (2014) Tangfeifania diversioriginum gen. nov., sp. nov., a representative of the family Draconibacteriaceae. Int J Syst Evol Microbiol 64:3473–3477
Meier-Kolthof JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genomebased taxonomy. Nat Commun 10:2182
Na SI, Kim YO, Yoon SH, Ha S, Baek I et al (2018) UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:280–285
Ou D, Huang H, Bai R, Li Q, Wang Y et al (2017) Nitratireductor aestuarii sp. nov., a marine alphaproteobacterium isolated from an estuary. Int J Syst Evol Microbiol 67:1637–1642
Pridham TG, Gottlieb D (1948) The utilization of carbon compounds by some Actinomycetales as an aid for species determination. J Bacteriol 56(1):107
Price MN, Dehal PS, Arkin AP (2010) FastTree 2 – Approximately Maximum-Likelihood trees for large alignments. PLOS ONE 5:e9490. https://doi.org/10.1371/journal.pone.0009490
Ramasamy D et al (2014) A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol 64:384–391
Rodriguez -RLM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4:e1900v1
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B (2003) The COG database:anupdated version includes eukaryotes. BMC Bioinformatics 41:4
Tindall BJ, Sikorski J, Smibert RM, Krieg NR (2007) Phenotypic characterization and the principles of comparative systematics. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf G, Schmidt TM et al (eds) Methods for General and Molecular Microbiology, 3rd edn. ASM Press, Washington, DC, pp 330–393
Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595
Xiao CL, Chen Y, Xie SQ, Chen KN, Wang et al (2017) MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat Methods 14:1072–1074
Yoon SH, Ha SM, Kwon S et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Yu Z, Zhuang L, Pan J, Wang Y, Zhou S (2016) Nitratireductor lacus sp. nov., isolated from Yuncheng salt lake, China. Int J Syst Evol Microbiol 66:4963–4967
Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA and Krieg NR (editors). Methods for General and Molecular Bacteriology
Acknowledgements
We would like to thank Editage (www.editage.cn) for English language editing.
Funding
This work was supported by the Stable Support to Agricultural Sci-Tech Renovation (xjnkywdzc-2022005), Regional Collaborative Innovation Special Project of Xinjiang Uygur Autonomous Region (Nos. 2021E02022) and Forestry Development Subsidy Fund Project of Xinjiang Uygur Autonomous Region (Nos. XJLYKJ-2021-15).
Author information
Authors and Affiliations
Contributions
XPY wrote the manuscript and analysed the cultivation data. LYZ and XWW performed the genomic and phylogenetic analysis. JPD and PBL isolated the strain and performed the initial cultivation and strain deposition. ZFW contributed to text preparation and revised the manuscript. ZHT took the samples. YQX corrected and reviewed the draft. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, X., Zhou, L., Wang, X. et al. Nitratireductor luteus sp. nov. isolated from saline-alkali land. Antonie van Leeuwenhoek 116, 221–229 (2023). https://doi.org/10.1007/s10482-022-01797-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-022-01797-7