Abstract
A Gram-stain-negative, non-motile and short-rod-shaped bacterium, designated as strain SY7T, was isolated from rhizosphere soil of the mangrove Kandelia obovata of Fugong village, in Zhangzhou, China. The isolate grew at 10–45 °C (optimum 30 °C), pH 6.0–10.0 (optimum pH 7.0) and 0–8% NaCl (optimum 3%, w/v). The 16S rRNA gene sequence and phylogenetic analysis revealed that strain SY7T located within the radiation of genus Nitratireductor and showed the highest sequence similarity of 97.23% to Nitratireductor pacificus MCCC 1A01024T. The DNA G+C content was 64.9%. In silico DNA–DNA hybridization and average nucleotide identity values between strain SY7T with reference strains of N. pacificus MCCC 1A01024T, N. basaltis KCTC 22119T and N. aquibiodomus DSM 15645T were 16.7%, 14.3%, 14.7% and 75.2%, 72.6%, 73.5%, respectively. The major isoprenoid quinone was Q-10. The dominant fatty acids were 11-methyl C18:1ω7c, iso-C17:0, C19:0ω8c cyclo and summed feature 8 (C18:1ω6c/C18:1ω7c), a profile that almost matched the other members of the genus Nitratireductor. The predominant polar lipids were phosphatidylcholine, phosphatidylglycerol, phosphatidylethanolamine, diphosphatidylglycerol. On the basis of the phenotypic, phylogenetic and chemotaxonomic analysis, strain SY7T represents a novel species of the genus Nitratireductor, for which the name Nitratireductor mangrovi sp. nov., is proposed. The type strain is SY7T (= KCTC 72110T = MCCC 1K03723T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Nitratireductor belongs to the family Phyllobacteriaceae of class Alphaproteobacteria and was originally described in 2004 by Labbé et al. [1]. At the time of writing, there are 8 species of the genus Nitratireductor with validly published names have been proposed according to LPSN (www.bacterio.net/nitratireductor.html). Most members of the genus Nitratireductor share some common features, such as being Gram-negative, catalase- and oxidase-positive, non-spore forming, rod shaped morphology, white-pigmented, nitrate reduction activity, rod shaped morphology, containing ubiquinone-10 (Q-10) as major isoprenoid quinone [1,2,3,4,5,6,7,8]. In this study, we described a bacterium strain SY7T, which isolated from a rhizosphere soil sample of mangrove forest. The isolate is considered to represent a novel species of the genus Nitratireductor, for which the name Nitratireductor mangrovi sp. nov., is proposed by using the polyphasic taxonomical approaches, including phenotypic, genotypic and chemotaxonomic analysis.
Materials and Methods
Isolation and Cultivation
Strain SY7T was isolated from a soil sample collected from rhizosphere soil of the mangrove Kandelia obovata of Fugong village (117° 57′ N 24° 24′ E), in Zhangzhou, China, and stored at 4 °C until use. Serially diluted (tenfold dilutions each) samples were made and spread on MA plates by the traditional dilution-plating method, and then incubated at 30 °C. After 15 days of incubation, a white-pigmented colony was picked and named as SY7T. After repeated plate streaking on the same medium, pure strains were obtained from individual colonies and preserved at − 80 °C as suspension with 25% (v/v) glycerol for further used [9]. Type strains N. pacificus MCCC 1A01024T, N. basaltis KCTC 22119T and N. aquibiodomus DSM 15645T were obtained from the Marine Culture Collection of China (MCCC), Korean Collection for Type Cultures (KCTC) and the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), repectively. All the strains were cultured under the same conditions for the experiments.
16S rRNA Gene Sequence and Phylogenetic Analysis
The 16S rRNA gene sequence of strain SY7T was amplificated with universal primers 27F [5′-AGAGTTTGATCCTGGCTCAG-3′] and 1492R [5′-ACGGCTACCTTGTTACGACTT-3′] by the method as described by Lane [10]. PCR products were ligated to vector pMD 19-T (TaKaRa) and cloned into E. coli DH5a for sequencing, and the almost-complete 16S rRNA gene sequence was obtained. The sequence was submitted to NCBI (https://www.blast.ncbi.nlm.nih.gov/Blast.cgi) (Accession No: MN239498) and compared with the closely related taxa provided by the EzTaxon-e server (https://eztaxon-e.ezbiocloud.net) [11, 12]. The multiple sequences were aligned with Clustal W [13]. Phylogenetic trees were reconstructed with MEGA 5.0 using neighbor-joining [14], maximum-parsimony [15] and maximum-likelihood [16] methods with Kimura two-parameter model. Bootstrap method (1000 resample datasets) was used to evaluate the trees topology [17, 18]. The type strains of N. pacificus MCCC 1A01024T, N. basaltis KCTC 22119T and N. aquibiodomus DSM 15645T were selected as reference strains.
Genome Sequence and Analysis
The genome sequence of strain SY7T was sequenced by Solexa PE150 sequencing technology with the HiSeq platform (Beijing Genomics Institute). The denovo assembly of the reads was performed using ABySS 1.5.2 [19]. The assembly k-value was tested from 32 to 64 to find the optimal k-value using abyss-pe script. The quality of microbial genome was assessed using Check M [20]. The open reading frames (ORFs) were predicted and annotated by Rapid Annotation using Subsystem Technology (RAST) server online [21]. The genomes of reference type strains of N. pacificus MCCC 1A01024T (AMRM00000000), N. basaltis KCTC 22119T (JMQM00000000) and N. aquibiodomus DSM 15645T (BAMP00000000) were downloaded from the NCBI database. In silico DNA–DNA hybridization (isDDH) values were calculated by genome to genome distance calculator (GGDC) [22]. The average nucleotide identity (ANI) values were calculated using the OrthoANIu algorithm of the Chun lab’s online Average Nucleotide Identity calculator [23]. The DNA G+C content of strain SY7T was calculated from the genome sequence.
Morphology, Physiology, Biochemistry and Chemotaxonomy
Cell morphology was determined by using optical microscopy (BX40; Olympus) and transmission electron microscopy (JEM-1230; JEOL) as described by Chen et al. [24]. Motility was tested by hanging drop method and semi-solid agar method [25]. The pH range for growth (pH 5–10, with intervals of 0.5 pH units) was investigated in MB medium by using the appropriate biological buffers: citrate/phosphate (pH 5.0 and 5.5), MOPS (Sigma) (pH 6.0–8.0), boric acid/borax (pH 8.5 and 9.0) and borax/NaOH (pH 9.5 and 10.0). The temperature range for growth was investigated at 4, 10, 15, 20, 25, 28, 30, 32, 35, 37, 40, 45 and 50 °C in MB medium. Growth at NaCl concentration range (0–12%, w/v, at intervals of 0.5%) was investigated in NaCl-free MB medium (according to MB formula, but without NaCl). Gram-staining was performed by the following method outlined by Claus et al. [26]. Oxidase and catalase activities were examined by the addition of 1% (w/v) tetramethyl-p-phenylene diamine and 3% (w/v) H2O2 solution, respectively [27]. Other physiological and biochemical activities tests were processed in API 20NE, API ZYM (bioMérieux) and GENIII MicroPlates (Biolog) according to the manufacturers’ instructions. Unless otherwise indicated, the tests of physiological and biochemical activities between strain SY7T and all reference strains were cultured in MB medium at 30 °C.
Whole-cell fatty acids of strain SY7T were analyzed according to Sasser M [28]. For the preparation of cellular fatty acid methyl esters (FAMEs), cells of strain SY7T, N. pacificus MCCC 1A01024T, N. basaltis KCTC 22119T and N. aquibiodomus DSM 15645T were harvested and lyophiled after cultured on MA for 3 days at 30 °C. Identification and quantification of the FAMEs were performed by the Sherlock Microbial Identification System (MIDI) with the standard MIS Library Generation software version 4.5 (Microbial ID) [29, 30]. Cells of strain SY7T cultured in MB medium for 5 days at 30 °C which used for respiratory quinones and polar lipids analysis. Isoprenoid quinones were extracted according to the procedure described by Minnikin et al. [31], and analyzed by using HPLC–MS (Agilent 1200 and Thermo Finnigan LCQ DECA XP MAX mass spectrometer) [32]. Polar lipids of the isolate were extracted from 3.0 g freeze-dried cells, then separated by two-dimensional TLC on silica gel 60F254 plates (Merck) and identified as previously described [31, 33, 34].
Results and Discussion
16S rRNA Gene Sequence and Phylogenetic Analysis
The PCR-based 16S rRNA gene sequence and genome-based 16S rRNA gene sequence were identical. And the 16S rRNA gene sequence (1411 bp) of strain SY7T indicated that the novel isolate belonged to the genus Nitratireductor and showed the highest sequence similarities to N. pacificus MCCC 1A01024T (97.23%), N. basaltis KCTC 22119T (96.83%) and N. aquibiodomus DSM 15645T (96.52%). In all phylogenetic trees (Fig. 1, Fig. S1), strain SY7T belonged to the cluster of the genus Nitratireductor and formed an independent lineage with moderate bootstrap support.
Neighbor-joining tree based on 16S rRNA gene sequences, showing the phylogenetic relationship between strain SY7T and other related species of the genus Nitratireductor. Brucella melitensis ATCC 23456 T, Rhizobium leguminosarum ATCC 10313 T and Hyphomicrobium denitrificans DSM 1869T were used as outgroup. Bootstrap values were expressed as a percentage of 1000 replicates and only those at or above 50% were given at the branch points. Bar, 0.01 substitutions per nucleotide position
Genome Sequence and Analysis
The draft genome sequence of strain SY7T generated 584 Mb of clean data. The genome completeness of strain SY7T was 98.72% with 0.82% contamination, which considered as good reference genome for deeper analysis (≥ 95% completeness, ≤ 5% contamination). The draft genome sequence of strains SY7T has a genome size of 4,838,603 bp and yielded 70 contigs after assembly. N50 value of strains SY7T was 217,092 bp with the largest contig of 346,613 bp. As a result, the quality of the genome was high enough for taxonomical analysis [35]. The genome in silico DNA–DNA hybridization (GGDC) values of strain SY7T and Nitratireductor pacificus MCCC 1A01024T, Nitratireductor basaltis KCTC 22119T and Nitratireductor aquibiodomus DSM 15645T were 16.7%, 14.3% and 14.7%, respectively. The average nucleotide identity (ANI) values of strain SY7T and Nitratireductor pacificus MCCC 1A01024T, Nitratireductor basaltis KCTC 22119T and Nitratireductor aquibiodomus DSM 15645T were 75.2%, 72.6% and 73.5%, respectively. These results were lower than the 70% threshold value for GGDC and 95–96% for ANI proposed for the delineation of bacterial species, which indicating that strain SY7T was distinguished from the reference strains of the genus Nitratireductor [35, 36]. The genomic DNA G+C content of strain SY7T was 64.9%, different from those of Nitratireductor pacificus MCCC 1A01024T (63 mol%) [2], Nitratireductor basaltis KCTC 22119T (56.7 mol%) [3], Nitratireductor aquibiodomus DSM 15645T (57 mol%) [1].
Morphology, Physiology, Biochemicstry and Chemotaxonomy
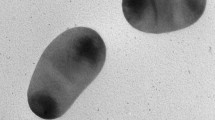
Strain SY7T formed smooth, circular, convex and creamy colonies with a diameter of 0.5–1.0 mm after 5 days of incubation at 30 °C on MA. The isolate was Gram-negative, positive for catalase, oxidase and nitrate reduction, which were in accordance with the characteristics of the genus Nitratireductor. Strain SY7T could be distinguished from its relatives by their phenotypic, physiological and biochemical characteristics. For example, cells of strain SY7T were short-rod-shaped without flagellum (Fig. 2), positive for hydrolysis of urea, assimilation of D-turanose, d-gluconic acid, negative for utilization of α-chymotrypsin, α-glucosidase, N-acetyl-glucosamine. Detailed characteristics between strain SY7T and the closely related species are shown in Table 1.
The major fatty acids of strain SY7T were 11-methyl C18:1ω7c (8.0%), iso-C17:0 (10.3%), C19:0ω8c cyclo (22.6%) and summed feature 8 (C18:1ω6c/C18:1ω7c) (22.4%). Detailed fatty acids profiles of strain SY7T and other closely related strains shown in Table 2. The fatty acids profiles of strain SY7T were similar to those of the reference strains, such as larger amounts of C19:0ω8c cyclo and summed feature 8 (C18:1ω6c/C18:1ω7c). But strain SY7T contained more iso-C17:0 (10.3%), 11-methyl C18:1ω7c (8.0%), iso-C15:0 (4.0%), anteiso-C15:0 (3.9%) and C8:0 3-OH (3.2%). Respiratory quinone analysis revealed that the sole quinone of strain SY7T was ubiquinone-10 (Q-10), same as the other species of the genus Nitratireductor [6,7,8]. The isolate contained phosphatidylcholine (PC), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), diphosphatidylglycerol (DPG) as major polar lipids, which was in agreement with data published previously for other species of the genus Nitratireductor [7, 8]. In addition, strain SY7 was also detected with an unidentified phospholipid (PL) and two unidentified lipid (L) (Fig. S2).
Taxonomic Conclusion
On the basis of the polyphasic taxonomical analysis as presented above, strain SY7T, which isolated from a rhizosphere soil sample of the mangrove forest, is considered to represent a novel species of the genus Nitratireductor, for which the name Nitratireductor mangrovi sp. nov., with strain SY7T as the type strain, is proposed.
Description of Nitratireductor mangrovi sp. nov.
Nitratireductor mangrovi (man.gro'vi. N.L. gen. n. mangrovi of a mangrove, referring to the isolation of the type strain from mangrove soil).
Cells are Gram-negative, non-motile, non-spore-forming, short-rod-shaped, approximately 0.4–0.7 μm in width and 1.0–1.5 μm in length. Colonies are 0.5–1.0 mm in diameter, smooth, circular, convex, creamy after incubation on MA for 5 days at 30 °C. The pH, temperature and NaCl concentration ranges for growth are determined to pH 6.0–10.0 (optimum pH 7.0), 10–45 °C (optimum 30 °C) and 0–8% NaCl (optimum 3.0%, w/v), respectively. Positive for oxidase, catalase, nitrate reduction, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, acid phosphatase and naphthol-AS-Bi-phosphatase, hydrolysis of esculin, β-galactosidase, urea and d-glucose, assimilation of d-maltose, d-trehalose, d-cellobiose, sucrose, N-acetyl-d-glucosamine, d-mannose, glycyl-l-proline, l-alanine, l-glutamic acid, l-pyroglutamic acid, l-lactic acid, citric acid, α-ketoglutaric acid, Tween 40, γ-amino-butyric acid, acetoacetic acid, acetic acid, d-turanose, α-d-glucose, d-fructose, d-gluconic acid, β-methyl-d-glucoside, quinic acid and dl-fucose, 1% sodium lactate, tetrazolium blue, lithium chloride, potassium tellurite and aztreonam. The predominant fatty acids are 11-methyl C18:1ω7c, iso-C17:0, C19:0ω8c cyclo and summed feature 8 (C18:1ω6c/C18:1ω7c). The isoprenoid quinone is Q-10. The major polar lipids are phosphatidylcholine (PC), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), diphosphatidylglycerol (DPG), and moderate amounts of an unidentified phospholipid (PL), two unidentified lipids (L) are also detected. The DNA G+C content of strain SY7T is 64.9%.
The type strain is SY7T (= KCTC 72110T = MCCC 1K03723T), and was isolated from rhizosphere soil of the mangrove Kandelia obovata of Fugong village in Zhangzhou, China. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain SY7T is MN239498. Whole Genome Shotgun project of strain SY7T has been deposited at DDBJ/ENA/GenBank under the accession number CP042301.
References
Labbé N, Parent S, Villemur R (2004) Nitratireductor aquibiodomus gen. nov. sp. nov., a novel alphaproteo bacterium from the marine denitrification system of the Montreal Biodome (Canada). Int J Syst Evol Microbiol 54:269–273
Lai Q, Yu Z, Yuan J et al (2011) Nitratireductor indicus sp. nov., isolated from deep-sea water. Int J Syst Evol Microbiol 61:295–298
Kim KH, Roh SW, Chang HW et al (2009) Nitratireductor basaltis sp. nov., isolated from black beach sand. Int J Syst Evol Microbiol 59:135–138
Lai Q, Yu Z, Wang J et al (2011) Nitratireductor pacificus sp. nov., isolated from a pyrene-degrading consortium. Int J Syst Evol Microbiol 61:1386–1391
Kang HS, Yang HL, Lee SD (2009) Nitratireductor kimnyeongensis sp. nov., isolated from seaweed. Int J Syst Evol Microbiol 59:1036–1039
Ou D, Huang H, Bai R et al (2017) Nitratireductor aestuarii sp. nov., a marine alphaproteo bacterium isolated from an estuary. Int J Syst Evol Microbiol 67:1637–1642
Yu Z, Zhuang L, Pan J et al (2016) Nitratireductor lacus sp. nov., isolated from Yuncheng Salt Lake. China. Int J Syst Evol Microbiol 66:4963–4967
Jang GI, Hwang CY, Cho BC (2011) Nitratireductor aquimarinus sp. nov., isolated from a culture of the diatom Skeletonemacostatum and emended description of the genus Nitratireductor. Int J Syst Evol Microbiol 61:2676–2681
Ye YY, Chen C, Ren YH et al (2019) Pseudomonas mangrovi sp. nov., isolated from mangrove soil. Int J Syst Evol Microbiol 69:377–383
Lane DJ (1991) 16S/23S rRNA sequencing. Nucleic Acid Tech Bact Syst 2:125–175
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Yoon SH, Ha SM, Kwon S et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. J Mol Biol Evol 4:406–425
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. J Syst Zool 20:406–416
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Tamura K, Peterson D, Peterson N et al (2011) MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. J Mol Biol Evol 28:2731–2739
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ et al (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) Check M: assessing the quality of microbial genomes recovered from isolates, single cells, and meta genomes. Genome Res 25:1043–1055
OverbeekR OR, Pusch GD, Olsen GJ, Davis JJ et al (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60–60
Lee I, Kim YO, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Micro Biol 66:1100–1103
Chen C, Anwar N, Wu C et al (2018) Halomonas endophytica sp. nov., isolated from liquid in the stems of Populuseuphratica. Int J Syst Evol Microbiol 68:1633–1638
Reddy CA (2007) Methods for general and molecular bacteriology. American Society for Microbiology, Washington DC
Claus D (1992) A standardized gram staining procedure. World J Microbiol Biotechnol 8:451–452
Cappuccino JG, Sherman N (2002) Microbiology: a laboratory manual, 6th edn. Benjamin Cummings, Menlo Park
Paisley R (1996) MIS whole cell fatty acid analysis by gas chromatography training manual. MIDI, Newark
Miller LT (1982) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol 16:584–586
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Inc., Newark
Minnikin DE, O'Donnell AG, Goodfellow M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Micro Biol Methods 2:233–241
Yu XY, Yu XD, Fu GY et al (2018) Marinicaulis flavus gen. nov. sp. nov., a novel stalked bacterium of the family Parvularculaceae. Int J Syst Evol Microbiol 68:2061–2067
Tindall BJ (1990) A comparative study of the lipid composition of Halobacterium saccharovorum, from various sources. Syst Appl Microbiol 13:128–130
Chen C, Su Y, Tao TY et al (2017) Maripseudobacter aurantiacus gen. nov. sp. nov., a novel genus of the family Flavobacteriaceae isolated from sedimentation basin. Int J Syst Evol Microbiol 67:778–783
Chun J, Oren A, Ventosa A et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. PNAS 106:19126–19131
Acknowledgement
This work was supported by grants from the Science and Technology Basic Resources Investigation Program of China (2017FY100300), the National Natural Science Foundation of China (No. 31700566) and the Siyuan Foundation (308000-521501/008).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequences of strain SY7T is MN239498. Whole Genome Shotgun project of strain SY7T has been deposited at DDBJ/ENA/GenBank under the accession CP042301.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ye, Y., Yan, C., Nie, Y. et al. Nitratireductor mangrovi sp. nov., a Nitrate-Reducing Bacterium Isolated from Mangrove Soil. Curr Microbiol 77, 1334–1340 (2020). https://doi.org/10.1007/s00284-020-01885-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01885-9