Abstract
A piezotolerant, cold-adapted, slightly halophilic bacterium, designated strain PWS21T, was isolated from a deep-sea sediment sample collected from the New Britain Trench. Cells were observed to be Gram-stain negative, rod-shaped, oxidase- and catalase-positive. Growth of the strain was observed at 4–45 °C (optimum 37 °C), at pH 5.0–9.0 (optimum 7.0) and in 0.5–20% (w/v) NaCl (optimum 3–4%). The optimum pressure for growth was 0.1 MPa (megapascal) with tolerance up to 70 MPa. 16S rRNA gene sequence analysis showed that strain PWS21T is closely related to Marinobacter guineae M3BT (98.4%) and Marinobacter lipolyticus SM19T (98.2%). Multilocus sequence analysis (MLSA) based on sequences of housekeeping genes gyrB, recA, atpD, rpoB and rpoD indicates that strain PWS21T represents a distinct evolutionary lineage within the genus Marinobacter. Furthermore, strain PWS21T showed low ANI and diDDH values to the closely related species. The principal fatty acids were identified as C12:0, C12:0 3-OH, C16:1ω9c, C16:0 and C18:1ω9c. Ubiquinone-9 was identified as the major respiratory quinone. The polar lipids were identified as phosphatidylethanolamine (PE), phosphatidylglycerol (PG), diphosphatidylglycerol (DPG), aminophospholipid (APL), two unidentified lipids and an unidentified phospholipid (PL). The G + C content of the genomic DNA was determined to be 60.3 mol%. On the basis of phenotypic, chemotaxonomic and molecular data, we conclude that strain PWS21T represents a novel species of the genus Marinobacter, for which the name Marinobacter profundi sp. nov. is proposed (type strain PWS21T = KCTC 52990T = MCCC 1K03345T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Marinobacter, belonging to the family Alteromonadaceae, was first proposed by Gauthier et al. (1992) and is comprised of 43 species isolated from diverse environments (Cui et al. 2016; Han et al. 2017). Species of this genus are Gram-stain negative, aerobic, and rod-shaped bacteria. The DNA G + C content of the genus ranges from 53.7 to 63.5 mol%, and the major fatty acids are C12:0, C16:1ω9c, C16:0 and C18:1ω9c, and the principal respiratory quinone is Q-9 (ubiquinone 9) (Gauthier et al. 1992; Cui et al. 2016; Han et al. 2017; Kim et al. 2017). The genus Marinobacter includes halophilic bacteria from marine-related environments and the type species of the genus, Marinobacter hydrocarbonoclasticus ATCC 49840T, is extremely halotolerant (Gauthier et al. 1992; Guo et al. 2007; Huo et al. 2008; Montes et al. 2008; Xu et al. 2008; Rani et al. 2017). During our recent screening of halophilic bacteria from samples collected from the New Britain Trench, we isolated bacterial strain PWS21T, which was able to grow in 20% NaCl. Strain PWS21T, isolated at a depth of 3908 m, was also found to be able to tolerate high hydrostatic pressure. The strain is phylogenetically related to members of the genus Marinobacter. In this study, characterization and classification of strain PWS21T were achieved using a polyphasic approach.
Materials and methods
Isolation and cultivation
Sediment samples were collected at a depth of 3908 m from the New Britain Trench (149.8°E, 6.6°S) in July 2016. Marine agar 2216 (MA; BD Difco) supplemented with NaCl (10%) was used to enrich the microbial consortium. Strain PWS21T was isolated by the standard dilution plating technique at 30 °C on MA. The routine cultivation of the strain and phenotypic tests were carried out on MA, unless noted otherwise. Strain PWS21T was maintained in marine broth 2216 (MB; BD Difco) with 20% (v/v) glycerol at − 80 °C. The type strains of Marinobacter guineae MCCC 1A00540T (= M3BT) and Marinobacter lipolyticus MCCC 1A03253T (= DSM 15157T = SM19T), provided by Marine Culture Collection of China (MCCC; Xiamen, China), were used as reference strains.
High pressure cultivation
High pressure growth experiments were performed in pressure vessels under a range of hydrostatic pressures (0.1–80 MPa) at 30 °C, supplemented with oxygen-saturated Fluorinert (FC-40, Sigma. 25% of total volume) to supply oxygen as previously described (Kato et al. 1995; Fang et al. 2006).
Phenotypic and chemotaxonomic analyses
Cellular morphology and flagellum formation were observed using atomic force microscopy (Multimode Nanoscope VIII; Bruker AXS) with cells grown on MA at 30 °C for 24 h (Su et al. 2012). Gram-staining, oxidase and catalase activity, Voges–Proskauer reaction, methyl red test, H2S production, indole production, and hydrolysis of starch, gelatin, casein, Tween 20, Tween 60 and Tween 80 were performed according to methods described by Dong and Cai (2001). The optimal growth temperature (4, 10, 20, 28, 37, 40, 45 and 50 °C) and pH (pH 4.0–11.0 in 0.5 unit increments, by using HOMOPIPES, MES, PIPES, HEPES and CAPS buffers, respectively), growth at different NaCl concentrations (0, 0.5, 1, 2, 3, 4, 5, 10, 15 and 20%, w/v) were determined as previously described (Lai et al. 2014). Anaerobic growth was evaluated in MB and in MB in the presence of NaNO3 (10 mM), prepared with a N2 gas phase (200 kPa) in sealed sterile vials and incubated at 30 °C for 7 days. Other biochemical tests were carried out using API 20NE, API ZYM and API 50CH strips (bioMérieux) according to the manufacturer’s instructions, with the NaCl concentration adjusted to 3.0% (w/v). Some tests in API strips, such as nitrate reduction, fermentation of d-glucose, aesculin hydrolysis and citrate utilization, were also re-examined by using conventional methods described by Dong and Cai (2001). To test carbon utilization, SO4PNsalts medium described by Cao et al. (2016), with a final concentration of 0.2% (w/v) carbon source was used.
Fatty acids in whole cells grown on MB at 30 °C for 48 h were saponified, extracted and methylated using the standard protocol of MIDI (Sherlock Microbial Identification System, version 6.0B). The fatty acids were analysed by gas chromatography (Agilent Technologies 6850) and identified using the TSBA6.0 database of the Microbial Identification System (Sasser 1990). The fatty acid profile of strain M. guineae MCCC 1A00540T (= M3BT) was performed in parallel with strain PWS21T under the same conditions (at the end of exponential growth phase). The respiratory quinone was extracted from cells of strain PWS21T and analysed by high pressure liquid chromatography (Agilent 1200 and Thermo Finnigan LCQ DECA XP MAX mass spectrometer) (Wu et al. 2015). Polar lipids were extracted from 100 mg of freeze-dried cellular material, separated by two-dimensional silica gel TLC (Merck) and then identified according to a previously described method (Tindall et al. 2007). The polar lipid profiles of strains M. guineae MCCC 1A00540T (= M3BT) and M. lipolyticus MCCC 1A03253T (= DSM 15157T = SM19T) were performed in parallel with strain PWS21T under the same conditions.
Phylogenetic analyses
Genomic DNA was prepared according to the method of Ausubel et al. (2002) and the 16S rRNA gene was amplified by PCR using primers Bac8F (5′-AGA GTT TGA TCA TGG CTC AG-3′) and U1492R (5′-GGT TAC CTT GTT ACG ACT T-3′) (Cao et al. 2016). Sequence similarity was determined using the EzBioCloud server (https://www.ezbiocloud.net/; Yoon et al. 2017). The phylogenetic analysis was performed using MEGA version 5.0 (Tamura et al. 2011). Distances were calculated using the Kimura two-parameter model and clustering was performed with the neighbor-joining (NJ) (Saitou and Nei 1987), maximum likelihood (ML) (Felsenstein 1981) and minimum evolution (ME) (Rzhetsky and Nei 1992, 1993) methods supported with bootstrap values based on 1000 replications. Multilocus Sequence Analysis (MLSA) based on sequences of housekeeping genes, gyrB (DNA gyrase β subunit), recA (recombinase A), atpD (ATP synthase β subunit), rpoB (RNA polymerase β subunit) and rpoD (RNA polymerase, sigma 70), was performed with the NJ, ML and ME methods supported with bootstrap values based on 1000 replications. Sequences of the housekeeping genes were downloaded from the NCBI database.
DNA–DNA relatedness
The draft genome sequences of strain PWS21T and M. guineae MCCC 1A00540T (= M3BT) were determined at Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China), using Solexa paired-end (500 bp library) sequencing technology. The de novo assembly of the reads was performed using SOAPdenovo 2.04 and GapCloser1.12 (Luo et al. 2012). The genome sequence of M. lipolyticus SM19T (ASAD00000000, Papke et al. 2013), M. gudaonensis CGMCC 1.6294T (GCA_900115175), M. salinus Hb8T (GCA_001854125), M. mobilis CN46T (GCA_900106945), M. adhaerens HP15T (GCA_000166295), M. segnicrescens SS011B1–4T (GCA_900111555), M. pelagius HS225T (GCA_900114925), M. hydrocarbonoclasticus ATCC 49840T (GCA_000284615) and M. salarius R9SW1T (GCA_000831005) were downloaded from the NCBI database. The average nucleotide identity (ANI) was calculated with the algorithm of Goris et al. (2007) using the EZGenome web service. The digital DNA–DNA hybridization (diDDH) estimate values were analysed using the genome-to-genome distance calculator (GGDC2.0) (Auch et al. 2010a, b; Meier-Kolthoff et al. 2013). The G + C content of the genomic DNA was determined from the draft genome sequence.
Results and discussion
High pressure growth
The optimum pressure for growth was determined to be 0.1 MPa, with tolerance up to 70 MPa (Fig. 1), which is much higher than that at the depth of origin (3908 m). This result shows that strain PWS21T is piezotolerant.
Phylogenetic analysis
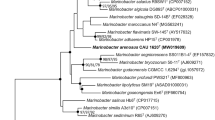
A nearly full-length 16S rRNA gene sequence (1409 nt, GenBank/EMBL/DDBJ accession MF800963) of strain PWS21T was determined. Comparative 16S rRNA gene sequence analysis showed that strain PWS21T formed a cluster within the genus Marinobacter, within the family Alteromonadaceae (Fig. 2). In all three trees (NJ, ME and ML trees), strain PWS21T formed a clade with M. guineae M3BT (Montes et al. 2008). Strain PWS21T shares high sequence similarity of 98.4% with M. guineae M3BT, followed by M. lipolyticus SM19T (98.2%) (Martin et al. 2003), M. goseongensis En6T (98.0%) (Roh et al. 2008), M. gudaonensis CGMCC 1.6294T (97.9%) (Gu et al. 2007), M. salinus Hb8T (97.5%) (Rani et al. 2017), M. mobilis CN46T (97.4%) (Huo et al. 2008), M. adhaerens HP15T (97.2%) (Kaeppel et al. 2012), M. segnicrescens SS011B1–4T (97.2%) (Guo et al. 2007), M. pelagius HS225T (97.2%) (Xu et al. 2008), M. hydrocarbonoclasticus ATCC 49840T (97.1%) (Gauthier et al. 1992), M. salarius R9SW1T (97.1%) (Ng et al. 2014) and M. maritimus CK47T (97.0%) (Shivaji et al. 2005). The level of 16S rRNA gene sequence similarities with closely related Marinobacter species showed that strain PWS21T displayed sufficient molecular differences for delineation at the species level, because the values fall well below the threshold value (98.65–98.70%) currently recommended for demarcation of distinct species (Stackebrandt and Ebers 2006; Kim et al. 2014; Chun et al. 2018). Further MLSA showed that strain PWS21T formed a cluster with the genus Marinobacter (Fig. 3) but represents a distinct evolutionary lineage within the genus Marinobacter. These results further affirmed that this strain represents a novel species.
Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences, showing the position of strain PWS21T and related species of the genus Marinobacter. Escherichia coli ATCC 11775T (X80725) was used as an outgroup. Filled circles indicate nodes that were also recovered in the maximum-likelihood and minimum evolution trees for the same sequences. Bootstrap values with neighbor-joining method (expressed as percentages of 1000 replications) are shown at branch points. Bar, 0.005 substitutions per nucleotide
MLSA of strain PWS21T and closely related species. Escherichia coli ATCC 11775T (JMST01000065) was used as an outgroup. The neighbor-joining tree was reconstructed based on five concatenated gene sequences (gyrB, recA, atpD, rpoB and rpoD). Filled circles indicate nodes that were also recovered in the maximum-likelihood and minimum evolution trees for the same sequences. Bootstrap values with neighbor-joining method (expressed as percentages of 1000 replications) are shown at branch points. Bar, 0.02 substitutions per nucleotide
The genome sequencing generated 1.04 and 1.17 G bytes clean data for strain PWS21T and M. guineae MCCC 1A00540T (= M3BT), respectively. The de novo assembly resulted in 39 contigs (strain PWS21T) and 37 contigs [M. guineae MCCC 1A00540T (= M3BT)]. The average genome coverage is 257 (strain PWS21T) and 261 (M. guineae MCCC 1A00540T). The draft genome accession numbers for strain PWS21T and M. guineae MCCC 1A00540T (= M3BT) are NTFH00000000 (4,034,600 bp with 39 contigs, N50 = 354,806 bp) and NTFI00000000 (4,458,854 bp with 37 contigs, N50 = 847,285 bp), respectively. The ANI values of strain PWS21T to M. guineae MCCC 1A00540T (= M3BT), M. lipolyticus SM19T, M. gudaonensis CGMCC 1.6294T, M. salinus Hb8T, M. mobilis CN46T, M. adhaerens HP15T, M. segnicrescens SS011B1–4T, M. pelagius HS225T, M. hydrocarbonoclasticus ATCC 49840T and M. salarius R9SW1T are 76.2%, 75.9%, 76.4%, 75.1%, 74.7%, 76.2%, 75.3%, 77.8%, 76.3% and 76.2%, respectively, which are below the standard ANI criterion for species identity (95–96%) (Richter and Rosselló-Móra 2009). The diDDH estimate values between strain PWS21T and M. guineae MCCC 1A00540T (= M3BT), M. lipolyticus SM19T, M. gudaonensis CGMCC 1.6294T, M. salinus Hb8T, M. mobilis CN46T, M. adhaerens HP15T, M. segnicrescens SS011B1–4T, M. pelagius HS225T, M. hydrocarbonoclasticus ATCC 49840T and M. salarius R9SW1T were 20.6 ± 2.5%, 20.3 ± 2.4%, 20.6 ± 2.5%, 20.3 ± 2.4%, 19.5 ± 2.4%, 20.7 ± 2.4%, 21.9 ± 2.4%, 21.7 ± 2.5%, 21.2 ± 2.5% and 20.7 ± 2.4%, respectively, which are far below the standard criterion (70%) for delineation of prokaryotic species (Wayne et al. 1987). These results confirm that strain PWS21T represents a novel genomic species of the genus Marinobacter.
Phenotypic characteristics
Cells of strain PWS21T were observed to be rod-shaped, approximately 1.2–2.3 μm long and 0.5–0.7 μm wide (Supplementary Fig. S1), catalase and oxidase positive, and Gram-stain negative. Colonies were observed to be smooth beige with regular edges on MA medium after 2 days of incubation at 30 °C. The strain was found to grow in 0.5–20% of NaCl (optimum 3–4%), from pH 5 to 9 (optimum 7), and at 4–45 °C (optimum 37 °C), but not at 50 °C within 2 weeks. Growth was observed under anaerobic conditions in the presence of NaNO3. Aesculin, Tween 80, Tween 60 and Tween 20 were observed to be hydrolysed, but gelatin, casin and starch were not. Voges–Proskauer reaction, methyl red test, glucose fermentation, indole production and H2S production was found to be negative. Nitrate reduction was observed to be positive. The strain was found to utilize d-glucose, lactose, adipic acid, pyruvate, malic acid, trisodium citrate and phenylacetic acid, but not formate, d-mannitol, N-acetyl-glucosamine, fumarate, d-mannose, d-maltose, d-fructose, potassium gluconate, l-arabinose, lactate, or dextrin. In the API ZYM tests, strain PWS21T was found to be positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, acid phosphatase, naphtol-AS-Bl-phosphohydrolase, and N-acetyl-β-glucosaminidase; negative for trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase, or α-fucosidase. In the API 20NE tests, strain PWS21T was found to utilize adipic acid, malic acid, trisodium citrate and phenylacetic acid, but not d-glucose, l-arabinose, d-mannose, d-mannitol, N-acetyl-glucosamine, d-maltose, potassium gluconate, or capric acid. Positive for reduction of nitrate, denitrification, beta-glucosidase (aesculin hydrolysis), but negative for indole production, d-glucose fermentation, gelatin hydrolysis or β-galactosidase. In the API 50CH tests, strain PWS21T was found to produce acid from glycerol, d-fructose, d-mannitol and d-glucose, but not others. The morphological, physiological and biochemical characteristics that differentiate strain PWS21T from closely related species are listed in Table 1.
Chemotaxonomic characteristics
The predominant fatty acids of strain PWS21T were identified as C12:0 (14.9%), C12:0 3-OH (19.4%), C16:1ω9c (18.6%), C16:0 (13.3%) and C18:1ω9c (15.5%) (Table S1). C12:0, C16:1ω9c, C16:0 and C18:1ω9c are characteristic of the genus Marinobacter (Cui et al. 2016), as well as strain PWS21T and M. guineae MCCC 1A00540T (= M3BT). These results indicate that strain PWS21T belongs to the genus Marinobacter, although several fatty acid features distinguished the novel strain from the closely related species, M. guineae MCCC 1A00540T (= M3BT). For example, a distinct feature was that M. guineae MCCC 1A00540T (= M3BT) contained considerable amounts of summed feature 3 (C16:1ω7c/C16:1ω6c, 17.4%), of which there was only 2.5% in strain PWS21T.
The polar lipids of strain PWS21T were identified as phosphatidylethanolamine (PE), phosphatidylglycerol (PG), diphosphatidylglycerol (DPG), aminophospholipid (APL), two unidentified lipids and an unidentified phospholipid (PL), as shown in Supplementary Fig. S2A. The polar lipid profile of strain PWS21T was similar to those of M. guineae MCCC 1A00540T (Fig. S2B) and M. lipolyticus MCCC 1A03253T (Fig. S2C), except some minor differences in unidentified lipids. The major respiratory quinone of strain PWS21T was identified as ubiquinone 9 (Q-9). This trait is in accordance with the properties of the genus Marinobacter. These results suggested that strain PWS21T is a member of the genus Marinobacter.
The DNA G + C content of strain PWS21T was determined to be 60.3 mol%, which is in the range previously reported for Marinobacter species (54.0–63.5 mol%) (Cui et al. 2016).
Strain PWS21T exhibits the typical characteristics of the genus Marinobacter. It has C12:0, C16:1ω9c, C16:0 and C18:1ω9c as the major fatty acids, Q-9 as the major respiratory quinone, and PE, PG, DPG, APL and PL as the major polar lipids. The differences in physiological, biochemical and chemotaxonomic characteristics of strain PWS21T and the closely species are given in Table 1. The data clearly shows differences between strain PWS21T and the reference strains. On the basis of these characteristics, together with their low ANI and diDDH values, we conclude that strain PWS21T represents a novel species of the genus Marinobacter, for which the name Marinobacter profundi sp. nov. is proposed.
The Digital Protologue database (Rosselló-Móra et al. 2017) TaxoNumber for strain PWS21T is TA00453.
Description of Marinobacter profundi sp. nov
Marinobacter profundi (pro.fun’di. L. gen. n. profundi of the depth of the ocean).
Cells are rods, about 1.2–2.3 μm long and 0.5–0.7 μm wide, catalase and oxidase positive, and Gram-stain-negative. On MA medium, produces smooth beige colonies with regular edges after 2 days of incubation at 30 °C. Growth occurs in 0.5–20% of NaCl (optimum 3–4%), from pH 5–9 (optimum 7), and at 4–45 °C (optimum 37 °C), but not at 50 °C within 2 weeks. The type strain is piezotolerant. The optimum pressure for growth is 0.1 MPa, with tolerance up to 70 MPa. Growth is observed under anaerobic conditions in the presence of NaNO3. Aesculin, Tween 80, Tween 60 and Tween 20 are hydrolysed, but gelatin, casin and starch are not. Nitrate reduction is positive, but Voges–Proskauer reaction, methyl red test, glucose fermentation, indole production and H2S production are negative. Principal fatty acids are C12:0, C12:0 3-OH, C16:1ω9c, C16:0 and C18:1ω9c. The major respiratory quinone is Q-9. The polar lipids comprise phosphatidylethanolamine (PE), phosphatidylglycerol (PG), diphosphatidylglycerol (DPG), aminophospholipid (APL), two unidentified lipids and an unidentified phospholipid (PL). The G + C content of the type strain is 60.3 mol%.
The type strain, PWS21T (= KCTC 52990T = MCCC 1K03345T), was isolated from a deep-sea sediment sample collected at a depth of 3908 m from the New Britain Trench, Solomon Sea (149.8°E, 6.6°S). The GenBank accession number for the 16S rRNA gene sequence of strain PWS21T is MF800963. The GenBank accession number for the genome sequence of strain PWS21T is NTFH00000000.
Abbreviations
- MCCC:
-
Marine Culture Collection of China
- KCTC:
-
Korean Collection for Type Cultures
- NCBI:
-
National Center for Biotechnology Information
- diDDH:
-
The digital DNA–DNA hybridization
- ANI:
-
The average nucleotide identity
References
Auch AF, Klenk HP, Göker M (2010a) Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2:142–148
Auch AF, von Jan M, Klenk HP, Göker M (2010b) Digital DNA–DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2:117–134
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman J, Smith JA, Struhl K (2002) Short protocols in molecular biology: a compendium of methods from current protocols in molecular biology. Wiley, New York
Cao J, Gayet N, Zeng X, Shao Z, Jebbar M, Alain K (2016) Pseudodesulfovibrio indicus gen. nov., sp. nov., a piezophilic sulfate-reducing bacterium from the Indian Ocean and reclassification of four species of the genus Desulfovibrio. Int J Syst Evol Microbiol 66:3904–3911
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Cui Z, Gao W, Xu G, Luan X, Li Q, Yin X, Huang D, Zheng L (2016) Marinobacter aromaticivorans sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from sea sediment. Int J Syst Evol Microbiol 66:353–359
Dong X, Cai M (2001) Determinative manual for routine bacteriology. Scientific Press, Beijing (English translation)
Fang J, Uhle M, Billmark K, Bartlett DH, Kato C (2006) Fractionation of carbon isotopes in biosynthesis of fatty acids by a piezophilic bacterium Moritella japonica strain DSK1. Geochim Cosmochim Acta 70:1753–1760
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Gauthier MJ, Lafay B, Christen R, Fernandez L, Acquaviva M, Bonin P, Bertrand JC (1992) Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int J Syst Bacteriol 42:568–576
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Gu J, Cai H, Yu SL, Qu R, Yin B, Guo YF, Zhao JY, Wu XL (2007) Marinobacter gudaonensis sp. nov., isolated from an oil-polluted saline soil in a Chinese oilfield. Int J Syst Evol Microbiol 57:250–254
Guo B, Gu J, Ye YG, Tang YQ, Kida K, Wu XL (2007) Marinobacter segnicrescens sp. nov., a moderate halophile isolated from benthic sediment of the South China Sea. Int J Syst Evol Microbiol 57:1970–1974
Han JR, Ling SK, Yu WN, Chen GJ, Du ZJ (2017) Marinobacter salexigens sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol 67:4595–4600
Huo YY, Wang CS, Yang JY, Wu M, Xu XW (2008) Marinobacter mobilis sp. nov. and Marinobacter zhejiangensis sp. nov., halophilic bacteria isolated from the East China Sea. Int J Syst Evol Microbiol 58:2885–2889
Kaeppel EC, Gardes A, Seebah S, Grossart HP, Ullrich MS (2012) Marinobacter adhaerens sp. nov., isolated from marine aggregates formed with the diatom Thalassiosira weissflogii. Int J Syst Evol Microbiol 62:124–128
Kato C, Sato T, Horikoshi K (1995) Isolation and properties of barophilic and barotolerant bacteria from deep-sea mud samples. Biodivers Conserv 4:1–9
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Kim JO, Lee HJ, Han SI, Whang KS (2017) Marinobacter halotolerans sp. nov., a halophilic bacterium isolated from a saltern crystallizing pond. Int J Syst Evol Microbiol 67:460–465
Lai Q, Cao J, Yuan J, Li F, Shao Z (2014) Celeribacter indicus sp. nov. a polycyclic aromatic hydrocarbon-degrading bacterium from deep-sea sediment and reclassification of Huaishuia halophila as Celeribacter halophilus comb. nov. Int J Syst Evol Microbiol 64:4160–4167
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18
Martin S, Marquez MC, Sanchez-Porro C, Mellado E, Arahal DR, Ventosa A (2003) Marinobacter lipolyticus sp. nov., a novel moderate halophile with lipolytic activity. Int J Syst Evol Microbiol 53:1383–1387
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60
Montes MJ, Bozal N, Mercade E (2008) Marinobacter guineae sp. nov., a novel moderately halophilic bacterium from an Antarctic environment. Int J Syst Evol Microbiol 58:1346–1349
Ng HJ, Lopez-Perez M, Webb HK, Gomez D, Sawabe T, Ryan J, Vyssotski M, Bizet C, Malherbe F, Mikhailov VV, Crawford RJ, Ivanova EP (2014) Marinobacter salarius sp. nov. and Marinobacter similis sp. nov., isolated from sea water. PLoS ONE 9:e106514
Papke RT, de la Haba RR, Infante-Dominguez C, Perez D, Sanchez-Porro C, Lapierre P, Ventosa A (2013) Draft genome sequence of the moderately halophilic bacterium Marinobacter lipolyticus strain SM19. Genome Announc 1:e00379-13
Rani S, Koh HW, Kim H, Rhee SK, Park SJ (2017) Marinobacter salinus sp. nov., a moderately halophilic bacterium isolated from a tidal flat environment. Int J Syst Evol Microbiol 67:205–211
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Roh SW, Quan ZX, Nam YD, Chang HW, Kim KH, Rhee SK, Oh HM, Jeon CO, Yoon JH, Bae JW (2008) Marinobacter goseongensis sp. nov., from seawater. Int J Syst Evol Microbiol 58:2866–2870
Rosselló-Móra R, Trujillo ME, Sutcliffe IC (2017) Introducing a digital protologue: a timely move towards a database-driven systematics of archaea and bacteria. Antonie Van Leeuwenhoek 110:455–456
Rzhetsky A, Nei M (1992) A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol 9:945–967
Rzhetsky A, Nei M (1993) Theoretical foundation of the minimum-evolution method of phylogenetic inference. Mol Biol Evol 10:1073–1095
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI, Newark
Shivaji S, Gupta P, Chaturvedi P, Suresh K, Delille D (2005) Marinobacter maritimus sp. nov., a psychrotolerant strain isolated from sea water off the subantarctic Kerguelen islands. Int J Syst Evol Microbiol 55:1453–1456
Stackebrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152
Su HN, Chen ZH, Liu SB, Qiao LP, Chen XL, He HL, Zhao X, Zhou BC, Zhang YZ (2012) Characterization of bacterial polysaccharide capsules and detection in the presence of deliquescent water by atomic force microscopy. Appl Environ Microbiol 78:3476–3479
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tindall B, Sikorski J, Smibert R, Krieg N (2007) Phenotypic characterization and the principles of comparative systematics. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf G, Schmidt TM, Snyder LR (eds) Methods for general and molecular microbiology, vol 3. ASM Press, Washington DC, pp 330–393
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wu YH, Xu L, Zhou P, Wang CS, Oren A, Xu XW (2015) Brevirhabdus pacifica gen. nov., sp. nov., isolated from deep-sea sediment in a hydrothermal vent field. Int J Syst Evol Microbiol 65:3645–3651
Xu XW, Wu YH, Wang CS, Yang JY, Oren A, Wu M (2008) Marinobacter pelagius sp. nov., a moderately halophilic bacterium. Int J Syst Evol Microbiol 58:637–640
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Acknowledgements
This work was supported by National Key R&D Program of China (Grant No. 2018YFC0310600) and by the National Natural Science Foundation of China (41773069, 41706146, 91328208, 41373071, and 41673085). The authors are grateful to all crews and on-board scientists of the M/V Zhangjian for taking the sediment samples in the New Britain Trench in September 2016.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cao, J., Liu, P., Liu, R. et al. Marinobacter profundi sp. nov., a slightly halophilic bacterium isolated from a deep-sea sediment sample of the New Britain Trench. Antonie van Leeuwenhoek 112, 425–434 (2019). https://doi.org/10.1007/s10482-018-1176-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-1176-8