Abstract
An elliptical to cucumber-shaped methanotroph with large cells was isolated from a rice rhizosphere in Western India. Strain Sn10-6 is one of the first methanotrophs to be isolated from Indian rice fields. Cells of Sn10-6 are Gram-negative, motile, large in size (3–6 µm × 1.5–2 µm) and contain intracellular cytoplasmic membrane stacks. Colonies of Sn10-6 and liquid cultures have a pale pink colour. Strain Sn10-6 was initially isolated under micro-oxic conditions but later adapted to grow under fully oxic conditions. The major fatty acids present were identified as C16:1ω6c, C16:1ω7c and C16:0 and ubiquinone was found to be the major quinone. The 16S rRNA gene sequence of strain Sn10-6 displays high similarity to the genes of Methylovulum psychrotolerans Sph1T (93.6%) followed by Methylosarcina fibrata AML-C10T (93.5%) and about 90–93% similarity to the genes of known species of Type I methanotrophic genera from the family Methylococcaceae. The draft genome information indicated that the G + C content of strain Sn10-6 is 43.9 mol%. Phylogenetic trees using 16S rRNA gene and the particulate methane mono-oxygenase sequences unequivocally placed Sn10-6 close to the genus Methylovulum. Based on the 16S rRNA gene differences, morphological characters and draft genome information, strain Sn10-6 (=MCC 3492 =KCTC 15683) is described here as the type strain of a novel species within a new genus, ‘Candidatus Methylocucumis oryzae’.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Methane is the second most important greenhouse gas after CO2 (https://www.epa.gov/gmi/importance-methane). Among the anthropogenic sources of methane, rice fields contribute to about 25.6 Tg CH4/year of the global methane emissions (Yan et al. 2009) which accounts for ~ 10% of global methane (Conrad 2009). India has the second largest area under rice cultivation in the world (Dubey 2005) contributing to about 3.9 Tg CH4/year (Ganesan et al. 2017). Methanotrophs living in the rhizospheres of rice plants are known to oxidise about 20% of the methane released from the rice ecosystems (Conrad 2009 and references therein).
Many bacterial and archaeal species remain undescribed as many of these microbes remain un-cultivated in the laboratory (Konstantinidis et al. 2017). Methanotrophs from rice fields have been mainly studied using cultivation independent approaches (Lüke 2010). Recently, a few new taxa of methanotrophs have been described from rice fields, which include the genera Methylogaea (Geymonat et al. 2011), Methylomagnum (Frindte et al. 2017; Khalifa et al. 2015) and Methyloterricola (Frindte et al. 2017). Previously known genera which have been isolated from rice fields include Methylomonas, Methylocaldum, Methylocystis and Methylosinus (Dianou et al. 2012; Ogiso et al. 2012). Within the family Methylococcaceae, which includes the Type Ia and Type Ib methanotrophs, seventeen genera have been reported (Bowman 2016; Frindte et al. 2017) (LPSN, http://www.bacterio.net/-classifphyla.html). Since only a very few isolates of methanotrophic bacteria from rice ecosystems are available for study, there is a need to isolate and characterise methanotrophs from this niche and study their properties.
A large sized gamma-proteobacterial methanotroph strain, Sn10-6, was isolated from the 10−6 dilution of a rhizosphere sample obtained from an Indian rice field (Pandit et al. 2016). This strain showed a unique 16S rRNA gene sequence with 93.6% or less similarity to validly described methanotrophic species indicating that it was a member of a putative novel genus and species (Pandit et al. 2016). The draft genome of strain Sn10-6 was sequenced to gain important insights into one of the first methanotrophic isolates from Indian rice fields (Rahalkar et al. 2016). In our present study, we formally characterise strain Sn10-6 by studying its morphological, metabolic and phylogenetic properties in detail and propose it to represent a novel genus and species within the Type I methanotrophs of the family Methylococcaceae.
Materials and methods
Isolation and cultivation of the strain
Strain Sn10-6 was isolated from the 10−6 dilution enrichment set up for isolation of methanotrophs using rice rhizosphere soil from Oryza sativa subsp. Indica (Indian var. Saal) from the Junnar disctrict (19°12′0″ North, 73°53′0″ East) of Western Maharashtra, India. The enrichment was set up in dilute nitrate mineral salts (dNMS) medium (Pandit et al. 2016) in a 48 well microtitre plate incubated in a desiccator with a headspace of ~ 20% methane, 25–30% air, and the remainder nitrogen, and incubated at 28 °C. After 4–5 weeks of incubation, culture suspension from wells showing turbidity or pellicle formation were streaked on the dNMS agarose (2%) plates and incubated again under similar conditions for 3–4 weeks. For isolation, individual colonies were picked and re-streaked repeatedly several times until the heterotrophs were completely removed. The purity was confirmed by no growth on 1:10 diluted nutrient agar plates. The pure colonies were then grown in dNMS liquid medium in glass flasks in desiccators or in sealed serum bottles with the same gas environment and incubated at 28 °C. Growth of strain Sn10-6 was also checked under variable oxygen concentrations (0, 5, 20 and 80% air, 20% methane and rest nitrogen) in serum bottles. For all experiments 10% inoculum of a freshly grown culture was used. For all of the physiological experiments and characterisation experiments, growth was performed in microtiter plates or flasks with liquid medium incubated under 80% air and 20% methane atmosphere at 25 °C, as maximum growth was seen under these conditions. Pure cultures were maintained by regular sub-culturing on dNMS plates and liquid medium and preserved at 4–8 °C (up to 4 months).
Morphological, cultural and physiological characteristics
For morphological examination the cells were observed under a phase contrast microscope (Nikon 80i, Japan microscope with a camera) with 40× and 100× magnifications. Cells were stained with DAPI (4′, 6-diamidino-2-phenylindole, 0.1 µg/ml final concentration) to observe the cell inclusions under fluorescence microscopy. Cells were also observed by Scanning Electron Microscopy (SEM) (Zeiss model EVO-MA-15 SEM). Cells were prepared in the following manner for SEM. A cell suspension of 0.2 OD was washed three times with phosphate buffered saline (PBS) followed by fixation in 2% glutaraldehyde and overnight incubation at 4 °C. Dehydration of the fixed cells with 500 µl ethanol (5%, 10%, and 20% up to 100%) was carried out by incubating the cell suspension at room temperature for 15 min followed by centrifugation at 12,000 rpm for 5 min at 4 °C, for each ethanol concentration. The final pellet was dissolved in a minimal volume of 100% ethanol and then mounted on black carbon taped stubs, coated with gold dust and observed under SEM to capture images and measure dimensions of the individual cells. Transmission Electron Microscopy (TEM) was also performed on the glutaraldehyde fixed cells of strain Sn10-6. The preparation of the sample was performed as described previously (Hoppert and Holzenburg 1998). Briefly, the cell pellet was initially suspended in molten agar (1.5%, w/v, final concentration). The resulting agar block was cut to small pieces of 1 mm3 volume and then the pieces were washed several times in distilled water. The pieces were then dehydrated in a graded ethanol series (15%, v/v, in dist. water, 30% for 15 min; 50%, 75%, 95% for 30 min; 100% for 1 h) at 0 °C. The samples were then incubated in LR White resin mixture (66%, v/v, in ethanol for 1 h; 100% for 10 h, 4 °C) and polymerised in pure resin for 24 h at 50 °C. Resin sections 80 nm in thickness were cut with glass knives or diamond knives in an ultramicrotome (Reichert Ultracut E, Leica Microsystems, Wetzlar, Germany) and stained with 4% (w/v) uranyl acetate solution (pH 7.0). Electron microscopy was performed in a Jeol JEM 1011 transmission electron microscope (Jeol Germany GmbH, Freising, Germany) at 80 kV and calibrated magnifications. Images were recorded with a Gatan Orius SC1000 CCD camera (Gatan, Munich, Germany). The TEM imaging was performed at University of Goettingen, Germany. Motility of the cells was observed by wet mount of a well grown cell suspension under 100× magnification. Gram character was determined by a standard Gram staining protocol.
Catalase activity of the culture was assessed by observing the production of bubbles after addition of 3% H2O2. Utilisation of various carbon sources was studied in dNMS liquid medium supplemented with the following filter-sterilised or autoclaved carbon substrates (0.1% w/v): formate, formamide, arabinose, raffinose, lactose, maltose, xylose, glucose, fructose and sucrose in microtitre plates. The ability of the strain Sn10-6 to grow on methanol was tested by the addition of 10–200 mM methanol in liquid medium in serum bottles. Similarly, the growth with formaldehyde (in the range of 1–50 mM) was also tested. The ability to utilise acetate as the sole carbon source was tested by supplying 2–10 mM range of sodium acetate in dNMS medium. The range of nitrogen sources utilised by strain Sn10-6 was tested by replacing KNO3 from dNMS medium with one of the following compounds: NH4Cl, urea, glycine, serine, valine, asparagine, aspartate, l-glutamic acid, glutamate, peptone or yeast extract. Growth without nitrogen source was tested under micro-oxic conditions (~ 4% oxygen).
The optimum pH and temperature ranges were evaluated in dNMS liquid medium. Growth at pH values in the range of 3–9.6 was checked after buffering the medium with citrate–phosphate buffer (pH 3–6.8) and glycine buffer (pH 8, 9 and 9.6). Growth was also checked without using any buffer, but using only HCl or NaOH for adjusting the pH from pH 3 to pH 10. Salt tolerance was tested with NaCl (0.1%, 0.5%, 1% and 2.0%, w/v) added to the mineral medium. All of the above experiments were performed in microtitre plates in triplicate. Strain Sn10-6 was grown at a temperature range from 10 to 40 °C, in liquid media in sealed serum bottles. The ability of the culture to grow under varying oxygen concentrations was tested in serum bottles by take in the range from no air to complete air in the headspace. The media preparation was done by flushing the bottles with nitrogen gas before autoclaving and then adding the calculated percentage of filtered air (0, 5, 2, 50 and 80%) with 20% methane in all bottles.
Chemotaxonomic analysis
Strain Sn10-6 was grown in 250 ml stoppered sealed bottles and a 10–12 days old culture, in the late exponential phase, was used for the analysis of cellular fatty acids. The culture was pelleted in 50 ml falcon tubes, pooled and a thick pellet was immediately transported on ice to the National Centre for Microbial Resources (NCMR) facility, Pune, India. There, the cells were subjected to saponification and methylation, followed by extraction, washes and injection of methyl esters for gas chromatography (7890A Agilent Technologies). The fatty acid methyl esters (FAME) profile obtained by gas chromatography was compared by the Sherlock pattern recognition software with a stored database for identification.
Phylogenetic analysis
DNA extraction from freshly grown pelleted cells was performed using the Gram-negative DNA extraction protocol of the Sigma GenElute™ Kit. The DNA concentration was determined using a Qubit Fluorometer 2.0. The 16S rRNA gene and pmoA gene amplification and sequencing for this isolate was achieved as described before (Pandit et al. 2016). This was routinely done after 6 months to confirm the identity of the maintained culture. Phylogenetic trees using the 16S rRNA gene and pmoA gene sequences from Sn10-6 and related species were constructed using the maximum likelihood algorithm based on the Tamura–Nei and Poisson correction, respectively, with the MEGA 6 software (Tamura et al. 2013) and 1000 bootstraps.
Information from draft genome
Genomic DNA was also used for whole genome shotgun sequencing using the Ion Torrent PGM (200 bp) platform; the assembly and annotation has been described elsewhere (Rahalkar et al. 2016). The contig data was submitted to the Rapid Annotation using Subsystem Technology (RAST) server (Aziz et al. 2008; Overbeek et al. 2014). BlastKOALA (http://www.kegg.jp/blastkoala/) and NCBI protein blast tools were used for the analysis of the draft genomes.
Digital DNA–DNA hybridization (dDDH, http://ggdc.dsmz.de/) (Meier-Kolthoff et al. 2013) was also performed using the draft genome of Sn10-6 and related species. A blast based pairwise average nucleotide identity ANI-b was calculated using JSpecies (Sangal et al. 2016; Richter and Rosselló-Móra 2009). Additionally, Ortho average nucleotide identity (ANI) calculations between Sn10-6 and closely related species was performed using the ORTHO ANI tool https://www.ezbiocloud.net/tools/orthoani (Yoon et al. 2017; Lee et al. 2016).
Results and discussion
Growth, morphological characteristics and microscopy of Sn10-6
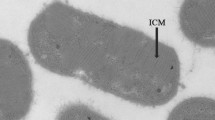
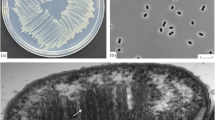
Growth was seen in the form of a pale pink pellicle in the 10−6 serial dilution well of the isolation plate. After the pellicle was streaked on dNMS agarose plates and incubated in a desiccator, pale pink coloured colonies appeared which were about 2–3 mm in diameter. When the cultures were viewed under the microscope, two types of cells were seen: numerous cells which were elliptical, large and fat rods which were motile; and thin rods, in small numbers. After several rounds of streaking from plate to plate, pure colonies of strain Sn10-6 were obtained on the plates. The colonies were observed to be light or pale pink in colour, round and measuring about 1 mm in diameter. The growth of pure colonies inoculated in dNMS liquid medium, after 6–7 days incubation, appeared as pale pink cell pellets settled to the bottom of the serum bottle, which on shaking formed a swirl of cells rising from the center of the bottom. Under phase contrast microscope, the cells were observed to be elliptical, large and fat rod shaped, resembling cucumbers as they had rounded ends, 3–4 × 1.5–1.8 µm (length × width) (Fig. 1a). The cells were found to be motile and Gram-stain negative. The dividing cells were observed to be elliptical (Fig. 1a). Chain formation was not often observed. In older cultures, Sn10-6 cells were determined to be relatively large size 5–6 µm × 1.5 μm. Strain Sn10-6 morphologically resembles strains of Methylomagnum ishizawaii, a rice field methanotroph species (Frindte et al. 2017). Characteristic inclusions or granules could be clearly observed by phase contrast microscopy (Fig. 1b). DAPI stained cells revealed the presence of fluorescent yellow stained intracellular granules (Fig. 1c), which are indicative of poly-phosphate granules. SEM images clearly showed the elliptical, cucumber shaped cells with an average cell size of 3.5 µm × 1.5 µm (Fig. 1d). Ultrathin sections also revealed the presence of ample storage granules throughout the entire cell (Fig. 1e). The draft genome of strain Sn10-6 contains genes for polyphosphate metabolism (WP_045778316.1) and genes related to the glycogen metabolism pathway (WP_082081464.1 and WP_082081754.1). On TEM strain Sn10-6 showed stacks of intracytoplasmic membranes arranged in stacks which were perpendicular to the cytoplasmic membranes mainly at the periphery of the cells (Fig. 1f), a typical characteristic of Type I methanotrophs (Whittenbury et al. 1970).
Phase contrast images of strain Sn10-6 (a–b), Cell morphology (a), unstained intracellular granules (b); UV fluorescence image of DAPI stained cells of strain Sn10-6 showing the presence of intracellular granules (c); SEM image of strain Sn10-6 showing clear cucumber shaped cells with marked cell size (d); TEM images of ultrathin sections of strain Sn10-6 (e–f) showing the intracellular granules (G)—(e) and membrane stacks (M) perpendicular to the cytoplasmic membrane (f)
Physiological characteristics and C and N metabolism
Strain Sn10-6 was found to utilise only methane or methanol as sole C sources. Cultures reached a maximum OD of 0.4 under completely oxic conditions, i.e. 80% air and 20% methane in the headspace. The culture can grow and tolerate methanol up to a concentration of 200 mM but did not grow above OD 0.28. Strain Sn10-6 failed to show any growth with other C sources such as formaldehyde, acetate, etc. for all the concentrations tested. Strain Sn10-6 can utilise ammonium chloride, l-serine, l-valine, peptone and yeast extract as sources of nitrogen.
Strain Sn10-6 was found to grow in the pH range of 5–9.6 under buffered as well as non-buffered conditions, with optimal growth at pH 6.5–7 (Table 1). Strain Sn10-6 was found to grow in the temperature range between 20 and 30 °C and to a limited extent at 37 °C. The optimum growth was observed at 25 °C, reflecting its mesophilic nature similar to other Type I genera (Table 1). The specific growth rates at 15 °C, 20 °C, 25 °C and 30 °C were 0.0006, 0.0023, 0.0038, 0.0019 h−1 and clumps were formed at 37 °C. No growth was observed at 10 °C or 40 °C. Strain Sn10-6 can tolerate NaCl concentrations up to 0.1% (w/v). Maximum growth in terms of OD (~ 0.4) was achieved at 80% air and 20% methane in serum bottles. Under 5% and 20% air atmosphere slow growth of strain Sn10-6 was observed and the OD measured was 0.18 after 7 days of incubation. The strain grows slowly and does not attain OD values higher than 0.4 and hence difficulties are faced in maintaining and preserving the strain. Additionally, the strain does not tolerate freezing conditions for long term preservation (− 80 °C in 15% glycerol for 4 months) but can be maintained by sub-culturing routinely or by storing at 4–8 °C for a period of 4 months.
The catalase activity test was negative. The draft genome of Sn10-6 also does not reveal the presence of catalase genes, but a peroxidase gene is present. The presence of ubiquinone as the major quinone was revealed by genome analysis of the strain Sn10-6 (Rahalkar et al. 2016).
Chemotaxonomic characteristics and phylogeny based on 16S rRNA gene and pmoA gene
Similar to other Type Ia methanotrophs, strain Sn10-6 was found to contain major amounts of C16 fatty acids (Table 2), with a predominance of C16:1ω6c/7c (summed feature; 58.2%) and C16:0 (20.2%). Members of the genus Methylovulum, the close phylogenetic relatives of strain Sn10-6, show a higher percentage of C16:0. Strain Sn10-6 was also found to contain fatty acids such as C14:0, C16:1ω11c, C16:1ω5c, C16:0 3OH and C18:0 in minor percentages (Table 2). Thus, Sn10-6 shows a distinctive FAME profile as compared to the other Type I methanotrophs.
The results of 16S rRNA gene sequence analysis using NCBI dis-continguous megablast (using sequences from type strains) showed that strain Sn10-6 is closely related to Methylovulum (Mvm.) psychrotolerans Sph1T (93.6%) and Methylosarcina (Msa.) fibrata AML-C10T (93.5%). A recently described strain of Mvm. psychrotolerans strain HV10-M2 (Mateos-Rivera et al. 2018) showed 93.8% 16S rRNA sequence similarity with Sn10-6 when all strains were included in the Blast analysis. The 16S rRNA gene similarities of Sn10-6 with other described Type I methanotrophs from the family Methylococcaceae were in the range of 90–93%. The 16S rRNA gene (KP793700.1) shows a large phylogenetic distance compared with other cultured Type Ia methanotrophs (6.2–10%). The current recommended 16S rRNA gene similarity cut-off for the demarcation of a new genus is ~ 94.5% or lower (Yarza et al. 2014), indicating that strain Sn10-6 belongs to a novel genus. Strain Sn10-6 showed a unique phylogenetic position within the phylogenetic trees constructed using both the 16S rRNA gene (Fig. 2) and the pmoA amino acid sequences of Type I methanotrophs (Fig. 3). It formed a distinct clade branching close to the genera Methylovulum, Methylosoma and Methyloglobulus in the 16S rRNA gene tree, within the Type I methanotrophs (Fig. 2). Mvm. psychrotolerans is a pink-pigmented large methanotroph reported from Northern peatlands (Oshkin et al. 2016) and recently from plant material from a high altitude environment (Mateos-Rivera et al. 2018). Based on the complete sequence of the pmoA gene, which codes for the beta subunit of particulate methane monooxygenase (pMMO), strain Sn10-6 shows close relatedness to Methylovulum miyakonese (Iguchi et al. 2011), i.e. 87% based on nucleotide sequence similarity. The amino acid sequence showed 94% sequence homology to pMMO of Mvm. miyakonese, followed by 92% homology with that of Mvm. psychrotolerans and showed more distant relationships to the sequences of other Type I methanotroph genera. In the pmoA-based tree, Sn10-6 grouped with the members of the genera Methylosoma and Methylovulum (Fig. 3).
Maximum-likelihood bootstrap tree of the 16S rRNA gene sequence of ‘Candidatus Methylocucumis oryzae’ Sn10-6 in comparison with those of other Type I methanotrophs. The evolutionary history was inferred by using the maximum-likelihood method based on the Tamura-Nei model and 1000 bootstraps. Bar represents 2% divergence
Maximum-likelihood bootstrap tree of the partial pMMO protein sequence of ‘Candidatus Methylocucumis oryzae’ Sn10-6 in comparison with partial pMMO proteins from other Type I methanotrophs. The evolutionary history was inferred by using the maximum-likelihood method based on the Poisson correction model and 1000 bootstraps. Bar represents 5% divergence
Draft genome information and salient genome characteristics
The draft genome (LAJX00000000.1; https://www.ncbi.nlm.nih.gov/genome/?term=Sn10-6) of strain Sn10-6 was analysed. The draft genome size is 4.58 Mb with a G + C content of 43.9% (Rahalkar et al. 2016). Strain Sn10-6 has a complete set of genes for the methane oxidation pathway, which includes the pMMO genes, methanol dehydrogenase, tetrahydromethanopterin pathway genes for formaldehyde oxidation and formate dehydrogenase genes (Rahalkar et al. 2016). Most of the annotated proteins in the draft genome sequence were most similar to those of Mvm. miyakonese (AQZU00000000.1) or Mvm. psychrotolerans HV10-M2 (CP022129) (Mateos-Rivera et al. 2018) or in some cases to other Type I methanotrophic species (Supplementary Table 1). Strain Sn10-6 also shows genes for the conversion of nitrite to nitric oxide and nitric oxide to nitrous oxide, and genes for nitrogen fixation, as reported recently in several Type I methanotrophs (Hamilton et al. 2015; Heylen et al. 2016; Kalyuzhnaya et al. 2015) (Supplementary Table 1). Although genes for nitrogen fixation are present we detected very limited growth on nitrogen free medium under micro-oxic conditions. The strain Sn10-6 genome contains three distinct copies of hemerythrin genes which have been recently discovered in many methanotrophic genomes and may be important for oxygen transport in methanotrophs (Rahalkar and Bahulikar 2018) (Supplementary Table 1).
For additional taxonomic information we performed dDDH between strain Sn10-6 and the closely related species, Mvm. psychrotolerans (CP022129) (Mateos-Rivera et al. 2018) and Msa. fibrata (ARCU00000000) (Hamilton et al. 2015). The dDDH value comparing Sn10-6 with Msa. fibrata was 16 ± 4%, whereas Mvm. psychrotolerans and Sn10-6 showed a dDDH value of 15 ± 2%, both of which are very low and support the conclusion that strain Sn10-6 belongs to a distinct species and possibly genus. ANI-b (genome) comparisons of Sn10-6 with Mvm. pscyhrotolerans and Msa. fibrata yielded values of 69.42% and 67.32%, respectively, which is well below the threshold of 75%, for separating genera (Sangal et al. 2016). Ortho-ANI comparisons (Yoon et al. 2017) of Sn10-6 with Mvm. pscyhrotolerans and Msa. fibrata yielded values of 70% and 68%, respectively.
Based on a 16S rRNA gene sequence difference of more than 6.2% (Yarza et al. 2014), low DDH value of ~ 15–20% with closely related species, a distinctive FAME profile and metabolic characters, we conclude that strain Sn10-6 belongs to a new genus and species and thus propose it to be the type strain of ‘Candidatus Methylocucumis oryzae’. Although strain Sn10-6 could not be preserved by glycerol stocks used for long term preservation and has to be sub-cultured continuously (or can be stored at 4–8 °C for 4 months), the strain has been deposited at National Centre for Microbial Resources, Pune, India culture collection (= MCC 3492) and in the Korean Collection for Type Cultures, Korea (=KCTC 15683). However, we propose the species with Candidatus status as difficulties in culturing the strain mean we have been unable to deposit the strain in additional culture collections. The Digital Protologue database (Rossello-Mora et al., 2017) TaxoNumber for Ca. Methylocucumis oryzae strain Sn10-6 is CA00040.
Description of ‘Candidatus Methylocucumis’
Methylocucumis (Me.thy.lo.cu’cu.mis. N.L. neut. n. methylum the methyl group; L. masc. n. cucumis cucumber; N.L. masc. n. Methylocucumis methyl utilising cucumber-shaped organism).
Obligate methanotroph, which can utilise methane or methanol as the sole source of C and energy. Uses pMMO for methane oxidation. Gram-stain negative, aerobic, motile. Divides by binary fission. Presence of typical intracellular stacked membranes as described for Type I methanotrophs. Belongs to the family Methylococcaceae. Phylogenetic and genomic characters place it close to genus Methylovulum. The type species is ‘Candidatus Methylocucumis oryzae’, which was isolated from a rice root rhizosphere.
Description of ‘Candidatus Methylocucumis oryzae’
Ca. Methylocucumis oryzae (o.ry’zae. L. fem. n. Oryza genus name of rice; L. gen. n. oryzae of rice, as the type strain was isolated from rice rhizosphere).
Cells are large in size with an average length of 3–4 µm that can grow up to 5–6 µm × 1.5–1.8 µm. Cells are pale-pink pigmented, cucumber-shaped (fat, rounded) rods. Growth is observed only in the presence of methane or methanol (10–200 mM). C1 compounds are assimilated via the ribulose monophosphate pathway (RuMP). Grows best in the mesophilic temperature range (20–30 °C) and at neutral pH (6.5–7). Cells are motile and genes for chemotaxis including flagella and Type IV pili are present in the type strain genome. The major cellular fatty acids are C16:1ω6c/7c and C16:0. The G + C content as determined from the draft genome of the type strain is 43.9 mol%.
The type strain Sn10-6 (=MCC 3492, =KCTC 15683) was isolated from a flooded rice field rhizosphere sample from Junnar, Maharashtra State, India. The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene and pmoA sequences of strain Sn10-6 are KP793700 (16S rRNA gene) and KT180167 (pmoA gene). The accession number of the draft genome sequence is NZ_LAJX00000000.1.
References
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75
Bowman John (2016) Methylococcaceae. In: Whitman W (ed) Bergey’s manual of systematics of archaea and bacteria. Wliey, New York
Conrad R (2009) The global methane cycle: recent advances in understanding the microbial processes involved. Environ Microbiol Rep 1:285–292
Dianou Day, Ueno Chihoko, Ogiso Takuya, Kimura Makoto, Asakawa Susumu (2012) Diversity of cultivable methane-oxidizing bacteria in microsites of a rice paddy field: investigation by cultivation method and fluorescence in situ hybridization (FISH). Microbes Environ 27:278–287
Dubey SK (2005) Microbial ecology of methane emission in rice agroecosystem: a review. Appl Ecol Environ Res 3:1–27
Frindte Katharina, Maarastawi Sarah A, Lipski Andre, Hamacher Joachim, Knief Claudia (2017) Characterization of the first rice paddy cluster I isolate, Methyloterricola oryzae gen. nov., sp. nov. and amended description of Methylomagnum ishizawai. Int J Syst Evol Microbiol 67:4507–4514
Ganesan A, Rigby M, Lunt MF, Parker RJ, Boesch H, Goulding N, Umezawa T, Zahn A, Chatterjee A, Prinn RG, Tiwari YK, van der Schoot Marcel, Krummel PB (2017) Atmospheric observations show accurate reporting and little growth in India’s methane emissions. Nat Commun 8:836
Geymonat E, Ferrando L, Tarlera SE (2011) Methylogaea oryzae gen. nov., sp. nov., a mesophilic methanotroph isolated from a rice paddy field. Int J Syst Evol Microbiol 61:2568–2572
Hamilton R, Kits KD, Ramonovskaya VA, Rozova ON, Yurimoto H, Iguchi H, Khmelenina VN, Sakai Y, Dunfield PF, Klotz MG, Knief C, den Camp HJMO, Jetten MSM, Bringel F, Vuilleumier S, Svenning Mette M, Shapiro N, Woyke T, Trotsenko YA, Stein LY, Kalyuzhnayaa MG (2015) Draft genomes of Gammaproteobacterial methanotrophs Isolated from Terrestrial ecosystems. Genome Announc 3:1–3
Heylen K, Vos De P, Vekeman B (2016) Draft genome sequences of eight obligate methane oxidizers occupying distinct niches based on their nitrogen metabolism. Genome Announc 4:1–2
Hoppert M, Holzenburg A (1998) Electron microscopy in microbiology. BIOS Scientific Publishers in association with the Royal Microscopical Society, Oxford
Iguchi H, Yurimoto H, Sakai Y (2011) Methylovulum miyakonense gen. nov., sp. nov., a type I methanotroph isolated from forest soil. Int J Syst Evol Microbiol 61:810–815
Kalyuzhnaya MG (2016a) Methylomicrobium. Bergey’s Manual of Systematics of archaea and bacteria. Wiley
Kalyuzhnaya MG (2016b) Methylosarcina. Bergey’s Manual of Systematics of archaea and bacteria. Wiley
Kalyuzhnaya Marina G, Lamb Andrew E, McTaggart Tami L, Oshkin Igor Y, Shapiro Nicole, Woyke Tanja, Chistoserdova Ludmila (2015) Draft genome sequences of Gammaproteobacterial methanotrophs isolated from lake Washington sediment. Genome Announc 3:1–3
Khalifa A, Lee CG, Ogiso T, Ueno C, Dianou D, Demachi T, Katayama A, Asakawa S (2015) Methylomagnum ishizawai gen. nov., sp. nov., a mesophilic type I methanotroph isolated from rice rhizosphere. Int J Syst Evol Microbiol 65:3527–3534
Konstantinidis K, Rosselló-Móra R, Amann R (2017) Uncultivated microbes in need of their own taxonomy. ISME J 11:2399–2406
Lee I, Kim YO, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Lüke C (2010) Molecular ecology and biogeography of methanotrophic bacteria in wetland rice fields. Max-Planck-Institut für terrestrische Mikrobiologie, Marburg
Mateos-Rivera Alejandro, Islam Tajul, Marshall Ian P G, Schreiber Lars, Øvreås Lise (2018) High-quality draft genome of the methanotroph Methylovulum psychrotolerans Str. HV10-M2 isolated from plant material at a high-altitude environment. Stand Genom Sci 13:1–8
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 60:1–14
Ogiso T, Ueno C, Dianou D, Huy TV, Katayama A, Kimura M, Asakawa S (2012) Methylomonas koyamae sp. nov., a type I methane-oxidizing bacterium from floodwater of a rice paddy field. Int J Syst Evol Microbiol 62:1832–1837
Oshkin IY, Belova SE, Danilova OV, Miroshnikov KK, Rijpstra WIC, Damsté JSS, Liesack W, Dedysh SN (2016) Methylovulum psychrotolerans a cold adapted methanotroph from low-temperature terrestrial environments and emended description of the genus Methylovulum. Int J Syst Evol Microbiol 66:2417–2423
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214
Pandit Pranitha S, Rahalkar Monali, Dhakephalkar Prashant, Ranade Dilip R, Pore Soham, Arora Preeti, Kapse Neelam (2016) Deciphering community structure of methanotrophs dwelling in rice rhizospheres of an Indian rice field using cultivation and cultivation independent approaches. Microb Ecol 71:634–644
Rahalkar Monali C, Bahulikar R (2018) Hemerythrins are widespread and conserved for methanotrophic guilds. Gene Rep 11:250–254
Rahalkar Monali C, Pandit Pranitha S, Dhakephalkar Prashant K, Pore Soham, Arora Preeti, Kapse Neelam (2016) Genome characteristics of a novel type I methanotroph ‘Sn10-6’ isolated from a flooded Indian rice field. Microb Ecol 71:519–523
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Rossello-Mora Ramon, Trujillo Martha E, Sutcliffe Iain C (2017) Introducing a digital protologue: a timely move towards a database-driven systematics of archaea and bacteria. Antonie Van Leeuwenhoek 110:455–456
Sangal Vartul, Goodfellow Michael, Jones Amanda L, Schwalbe Edward C, Blom Jochen, Hoskisson Paul A, Sutcliffe Iain C (2016) Next-generation systematics: an innovative approach to resolve the structure of complex prokaryotic taxa. Sci Rep 6:1–12
Schink B, Rahalkar MC (2016) Genus Methylosoma Rahalkar, Bussmann and Schink 2007, 1078VP. In: Whitman W (ed) Bergey’s manual of systematics of archaea and bacteria. Wliey, New York
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Whittenbury R, Phillips KC, Wilkinson JF (1970) Enrichment, isolation and some properties of methane utilising bacteria. J Gen Microbiol 61:205–218
Yan X, Akiyama H, Yagi K, Akimoto H (2009) Global estimations of the inventory and mitigation potential of methane emissions from rice cultivation conducted using the 2006 intergovernmental panel on climate change guidelines. Glob Biogeochem Cycles 23
Yarza Pablo, Yilmaz Pelin, Pruesse Elmar, Glöckner Frank Oliver, Ludwig Wolfgang, Schleifer Karl-Heinz, Whitman William B, Euzéby Jean, Amann Rudolf, Rosselló-Móra Ramon (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645
Yoon SH, Ha SM, Lim JM, Kwon SJ, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Acknowledgements
The work was supported by funds from DBT BioCARe, India and institutional funds received from MACS Agharkar Research Institute, India. We acknowledge the farmer, Mr. Rengde who supported the rice field sample collection. We also acknowledge Ms. Aditi Purandare, Ms. Vishakha Kulkarni and Dr. Neelima Kulkarni for their help during collection of the sample. We acknowledge Dr. Prashant K. Dhakephalkar for his support during the project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pandit, P.S., Hoppert, M. & Rahalkar, M.C. Description of ‘Candidatus Methylocucumis oryzae’, a novel Type I methanotroph with large cells and pale pink colour, isolated from an Indian rice field. Antonie van Leeuwenhoek 111, 2473–2484 (2018). https://doi.org/10.1007/s10482-018-1136-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-1136-3