Abstract
From comparisons of ITS1-5.8S-ITS2 and gene sequences for nuclear D1/D2 LSU rRNA, nuclear SSU (18S) rRNA, translation elongation factor 1-α (EF1-α) and RNA polymerase II subunit 2 (RPB2), the following four new ascosporogenous yeast species were resolved and are described as Metschnikowia anglica (NRRL Y-7298T [type strain], CBS 15342, MycoBank MB 823167), Metschnikowia leonuri (NRRL Y-6546T, CBS 15341, MB 823166), Metschnikowia peoriensis (NRRL Y-5942T, CBS 15345, MB 823164) and Metschnikowia rubicola (NRRL Y-6064T, CBS 15344, MB 823165). The following six species of Candida are members of the Metschnikowia clade and are proposed for transfer to Metschnikowia as new combinations: Candida chrysomelidarum (NRRL Y-27749T, CBS 9904, MB 823223), Candida gelsemii (NRRL Y-48212T, CBS 10509, MB 823192), Candida kofuensis (NRRL Y-27226T, CBS 8058, MB 823195), Candida picachoensis (NRRL Y-27607T, CBS 9804, MB 823197), Candida pimensis (NRRL Y-27619T, CBS 9805, MB 823205) and Candida rancensis (NRRL Y-48702T, CBS 8174, MB 823224). Candida fructus (NRRL Y-17072T, CBS 6380, MB 823206) is transferred to Clavispora as a new combination, and Candida musae is shown to be a synonym of C. fructus. Apparent multiple alleles for ITS, D1/D2, EF1-α and RPB2 were detected in strains of some species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ascomycete yeast genus Metschnikowia was described more than 100 years ago (Kamienski 1899) and most of the species produce distinctive long, needle-shaped ascospores. Species are common worldwide and often isolated from flowers and insects but some, such as members of the Metschnikowia bicuspidata clade, are known to parasitize brine shrimp (Artemia spp.), Daphnia species and copepods (Lachance 2011, 2016 and references therein). The Metschnikowia pulcherrima clade is of particular interest to agriculture because Metschnikowia pulcherrima and Metschnikowia. fructicola are effective as biocontrol agents to inhibit fruit storage rots such as those caused by species of Penicillium, Botrytis and certain other fungi (Janisiewicz et al. 2001; Kurtzman and Droby 2001; Piano et al. 1997; Türkel et al. 2014). The mechanism of biocontrol is not entirely clear, but may be partially based on competitive inhibition through sequestration of iron via production of the iron-binding compound pulcherrimin (Oro et al. 2014; Sipiczki 2006). Additionally, M. fructicola produces chitinase enzymes in the presence of fungal pathogens (Banani et al. 2015; Wang et al. 2017) and these may assist in biocontrol. Although important for agricultural uses, few biotechnological applications have so far been demonstrated for species of Metschnikowia, although one example is stereoconversion of secondary alcohols by Metschnikowia koreensis (Meena et al. 2014).
The taxonomy of Metschnikowia has been partially clarified by DNA sequence comparisons, which have resulted in better definition of earlier described species recognized from phenotype and the discovery of numerous new species. Lachance (2016) reported that 81 species are now known for the Metschnikowia clade, but this clade is recognized from short DNA sequences, such as the D1/D2 domains of the LSU rRNA gene, or from concatenation of a few gene sequences. Consequently, branch support in phylogenetic trees based on these analyses is often weak and species relationships are uncertain. Examples of this problem are shown in the D1/D2 analysis of Lachance et al. (2011), the multilocus analysis of Guzmán et al. (2013) and the analysis given in the present study. However, even with weak branch support, it appears that Metschnikowia, as presently recognized, may actually represent several genera. The large-spored species, such as Metschnikowia aberdeeniae and Metschnikowia hawaiiensis, form a separate clade, the core species of the Clavispora clade are separate from neighbors and several nearby species groupings are potential genera. From this, it appears that the taxonomy of this large group of species can only be resolved from whole genome comparisons.
In the present work, four new ascosporogenous species are described and assigned to Metschnikowia and seven new combinations are proposed, six Candida species in Metschnikowia and one Candida species in Clavispora. These new species and new combinations are closely related to the type species of their respective genera and are expected to be unaffected by any future recircumscription of Metschnikowia and Clavispora.
Materials and methods
Isolation and phenotypic characterization of strains of the new species to be described
Most strains of the proposed new species examined in this study were isolated over many years by L. J. Wickerham, tentatively identified from phenotype as Metschnikowia pulcherrima or M. reukaufii, and accessioned into the ARS Culture Collection for future study. Isolation typically consisted of placing a substrate sample into a flask of liquid isolation medium that contained Yeast Nitrogen Base along with glucose, cellobiose, d-xylose, l-rhamnose, erythritol, d-mannitol and calcium 2-keto-gluconate and incubating at 25 °C for 1–2 weeks on a shaker. A loopful of growth from the flask was streaked on YM agar plates from which individual colonies were isolated (Wickerham 1969).
Fermentation and growth tests for strain characterization were conducted in liquid media following standard methods (Kurtzman et al. 2011) and incubated for at least 4 weeks at 25 °C. Individual strains and strain mixtures were tested for ascosporulation on some or all of the following media: YM agar (3 g yeast extract, 3 g malt extract, 5 g peptone, 10 g glucose, 20 g agar/l H2O); 5% malt extract agar (50 g malt extract, 30 g agar/l H2O); RG agar (0.2 g yeast extract, 0.2 g peptone, 1.0 g glucose and 20 g agar/l H2O); YCBAS agar (11.7 g Difco Yeast Carbon Base, 100 mg (NH4)2SO4, 20 g agar/l H2O), and 1:19 V8 juice agar (equal volumes of deionized water and V8 juice, adjust pH to 5.5 with NaOH, dilute 1:19 with water, agar to 2%, autoclave). Incubation times and temperatures are reported in the new species descriptions.
DNA isolation, sequencing and phylogenetic analysis
Methods for DNA isolation and sequencing of the internal transcribed spacer (ITS) and genes for domains D1/D2 of the nuclear large subunit rRNA (D1/D2), nuclear small subunit rRNA (18S), translation elongation factor-1α (EF-1α) and RNA polymerase II, subunit 2 (RPB2) were previously reported (Kurtzman and Robnett 1998, 2003). The most commonly used primers were: ITS, NS-7A and ITS4; 18S, NS-1AF, NS-3AF, NS-3AR, NS-5.4R, NS-7BR, NS-8R; D1/D2, NL-1, NL-4; EF-1α, YTEF-1F, YTEF-6AR, EF1-1577F, EF1-1567R, EF1-983F and EF-2218R; RPB2, YRPB2-1F, YRPB2-2F, YRPB2-2.2F, YRPB2-4F, YRPB2-5F, YRPB2-5R, YRPB2-6F, RPB2-6FR, RPB2-7R, YRPB2-8F, YRPB2-8R, YRPB2-9R, YRPB2-9.3R and YRPB2-11R. For phylogenetic analysis, sequences were aligned using Muscle, which is in the MEGA version 7.0.14 software package (Kumar et al. 2016), and the alignments were visually adjusted. Phylogenetic relatedness among species was determined from the maximum likelihood program included in MEGA 7.0.14 using the Hasegawa–Kishino–Yano model, and bootstrap support was determined from 1000 replicates.
Results and discussion
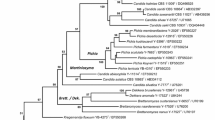
Relatedness among species of the “small-spored” members of Metschnikowia and the many Candida species that appear associated with them (Daniel et al. 2014; Guzmán et al. 2013; Kurtzman and Robnett 2013; Lachance 2011, 2016; Lachance et al. 2011) was assessed from maximum likelihood analysis of concatenated gene sequences for D1/D2 and EF-1α, and typical of analyses that include few gene sequences, deep nodes in the phylogenetic tree showed weak branch support (Fig. 1). The clade delimited by Metschnikowia sinensis and M. lachancei, which includes M. bicuspidata, type species of the genus Metschnikowia, was strongly supported (92% bootstrap support). Placement of certain other species, such as M. saccharicola and M. drosophilae, which are almost certainly members of Metschnikowia, are only weakly supported and continued assignment to the genus will require verification from more robust datasets, such as whole genome sequences.
Phylogenetic relationships among species of Metschnikowia, Clavispora and related clades determined from maximum likelihood analysis of concatenated gene sequences for D1/D2 LSU rRNA and EF-1α. GenBank accession numbers following strain numbers are D1/D2 and EF-1α, respectively. Bootstrap values > 50% are given at nodes and based on 1000 replicates. Schizosaccharomyces pombe was the designated outgroup species. Numbers of ambiguous nucleotides recorded for each sequences (D1/D2, EF-1α) are given in parentheses. Taxa in bold font represent new species and new combinations proposed in this study
The new species proposed for Metschnikowia in the current study are members of the M. bicuspidata clade, as are six Candida species proposed for transfer to Metschnikowia. An additional species, Candida fructus, is closely related to Clavispora lusitaniae, type species of Clavispora, and will be transferred to that genus. From the analysis presented in Fig. 1 and that of Guzmán et al. (2013), Metschnikowia and Clavispora appear to be separate genera. Multiple alleles were detected for certain genes in some species, and this will be discussed relative to species circumscription.
Description of new species of Metschnikowia
Metschnikowia peoriensis C. P. Kurtzman, C. J. Robnett & E. Basehoar sp. nov
Cell morphology
After 2 days on YM agar at 25 °C, cell morphology ranges from spherical (3–8 µm) to ellipsoidal (2–3 × 3–8 µm). Cell division is by multilateral budding and cells occur singly and in budded pairs (Fig. 2a). After 7 days at 25 °C, poorly differentiated pseudohyphae formed under the coverglass on a Dalmau plate with corn meal agar (Fig. 2b), but true (septate) hyphae were not detected. Colony growth is white, semi-glistening, butyrous in texture and low with a slightly irregular margin. Strains did not form pulcherrimin on either YM agar or V8 juice agar.
Metschnikowia peoriensis, NRRL Y-5942T. a Budding cells and a chlamydospore (arrow), YM agar, 2 days, 25 °C. b Simple pseudohypha, Dalmau plate, cornmeal agar, 7 days, 25 °C. c NRRL Y-5931 × NRRL Y-5934. Conjugating cells (arrows), 1:19 V8 juice agar, 12 days, 15 °C. d NRRL Y-5942. Asci with ascospores, 1:19 V8 juice agar, 9 days, 15 °C. Bar, 10 µm, all photographs
Ascosporic state
Eleven of the 18 known strains of M. peoriensis form ascospores, six represent mating types and one that has not been observed to form ascospores or to conjugate with either mating type (Table 1). Ascosporulation was observed on 1:19 V8 juice agar after incubation at 15 °C. NRRL Y-5942 and several other strains formed abundant asci with ascospores after 7 days, but other strains formed asci only after 4 weeks. Asci are unconjugated and develop from the large “pulcherrima” cells or “chlamydospores” that are abundantly present and assume a sphaeropedunculate shape (Fig. 2d). Asci measure 25–36 µm in length with the basal portion having a diameter of 5–8 µm. Asci are persistent and produce two closely paired acicular ascospores. Conjugation between mating types was tested on 1:19 V8 juice agar with incubation at 15 °C. Conjugants were usually detected within 2–3 days following mixing of complementary strains (Fig. 2c). Cultures with conjugants did not form asci, but when these cultures were transferred to YM agar, incubated for 3 days at 25 °C, and then transferred back to V8 agar and incubated at 15 °C, a limited number of asci with ascospores formed. Apparently, growth on YM agar replenished cellular nutrient levels. The most active mating pair was NRRL Y-5931 (a) and NRRL Y-5934 (α).
Fermentation and growth reactions for this species are given in Table 2.
Type
NRRL Y-5942 is designated as the type strain of Metschnikowia peoriensis. The strain is preserved as a metabolically inactive lyophilized preparation (the holotype) at the ARS Culture Collection, National Center for Agricultural Utilization Research, Peoria, IL, USA, and an isotype is preserved at the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands as CBS 15345. MycoBank number: MB 823164. Complementary mating types are NRRL Y-5931 (CBS 15336), mating type a, and NRRL Y-5934 (CBS 15337), mating type α. NRRL Y-5942, an ascosporogenous isolate, was isolated from the flower of a purple aster (Aster novi-belgii) collected in Peoria, Illinois, USA. Sources of other strains are given in Table 1.
Etymology
The species epithet peoriensis pertains to Peoria, Illinois, USA, the location that provided most of the strains examined in this study.
Ecology
Metschnikowia peoriensis is predominantly known from flowers of red clover (Trifolium pretense) and purple aster (Aster novi-belgii) collected in Peoria, Illinois, USA, and from two isolates obtained from wild blackberries (Rubus sp.) collected in Michigan, USA (Table 1). Because of the association with flowers, the species may be insect disseminated.
Relatedness to other Metschnikowia species
On the basis of maximum likelihood analysis of concatenated gene sequences of D1/D2 and EF-1α, M. peoriensis is most closely related to M. koreensis followed by M. cibodasensis, M. reukaufii and M. persici (Fig. 1). M. peoriensis differs from M. koreensis by substitutions in D1/D2 (6), EF-1α (6) and RPB2 (64) (Table 1). For these three sequences, there are essentially no differences between strains of M. peoriensis (Table 1), and unresolved nucleotides in the sequences for EF-1α and RPB2 usually occur in the same position for all strains. In contrast, the ITS1-5.8S-ITS2 sequence shows, for some strains, as many as 11 differences and all or nearly all occur in ITS1 (Table 1, Fig. 3). An examination of amplicons generated with primers NS-7A and ITS-4 shows two bands for the strains and because nearly all of the nucleotides can be resolved, it appears that the more concentrated amplicon determines the observed sequence. The forward primer Y5942-ITS1 (5′ CTCTAAATATTATATCTTC) was synthesized to match the NRRL Y-5942 ITS1 variable region and when used to generate an amplicon with primer ITS-4, still resulted in generation of two bands (Fig. 3). From this, it is concluded that more than two ITS1 alleles are present and resolution will only come from cloning this region of DNA. The occurrence of unresolved nucleotides is consistent with this observation. Resolution of the ITS1 sequence heterogeneity may be helped by an examination of NRRL Y-5957 and NRRL Y-5958. According to ARS Culture Collection records, these strains were derived as individual colonies from a streak plate of NRRL Y-5941. We have demonstrated that the strains represent complementary mating types and that NRRL Y-5957 has the substituted sequence shown by NRRL Y-5941, whereas the sequence of NRRL Y-5958 differs and matches that of the type strain, which we term “unsubstituted”. Despite ITS heterogeneity, little or no intraspecific variability was shown for the other genes sequenced and the 18 strains assigned to M. peoriensis appear to be members of the same species.
Amplicons of ITS1-5.8S-ITS2 from four strains of Metschnikowia peoriensis using primers Y5942-ITS1 and ITS4. Each amplification produced two bands, typical of amplifications using primers NS-7A and ITS-4. Primer Y5942-ITS1 was designed to match the ITS1 sequence of NRRL Y-5942 with the expectation that NRRL Y-5942, NRRL Y-5946 and NRRL Y-6044 would only produce single bands and that NRRL Y-5941 would produce no amplicons because of its many differences with the NRRL Y-5942 sequence. See text for discussion
Phenotypic differences are not sufficient to resolve M. peoriensis from other species of Metschnikowia. The key to species provided by Lachance (2011) places M. peoriensis among members of the M. pulcherrima clade, which are unresolved, demonstrating that identification of species must rely on comparison of DNA sequences.
Metschnikowia rubicola C. P. Kurtzman, C. J. Robnett & E. Basehoar sp. nov
Cell morphology
After 2 days on YM agar at 25 °C, cell morphology ranges from ellipsoidal (3–5 × 4–10 µm) to elongate (2–3 × 5–14 µm). Cell division is by multilateral budding and cells occur singly and in budded pairs (Fig. 4a). After 7 days at 25 °C, simple pseudohyphae were formed under the coverglass on a Dalmau plate with corn meal agar (Fig. 4b), but true (septate) hyphae were not detected. Colony growth is low, white, semi-glistening, butyrous in texture and with a smooth, slightly irregular margin. Five strains formed pulcherrimin on either YM agar or V8 juice agar.
Metschnikowia rubicola, NRRL Y-6064T. a Budding cells, YM agar, 2 days, 25 °C. b Simple pseudohypha, Dalmau plate, cornmeal agar, 7 days, 25 °C. c NRRL Y-6148 × NRRL Y-6066, conjugating cells (arrows), 1:19 V8 juice agar, 4 weeks, 15 °C. d NRRL Y-6064, ascus with an ascospore, 1:19 V8 juice agar, 12 weeks, 15 °C. Bar, 10 µm, all photographs
Ascosporic state
Seventeen strains of M. rubicola were examined in this study and of these, only two formed ascospores (Table 3). Seven of the strains represent mating types and the remaining eight strains gave ambiguous mating responses. Ascosporulation was observed on 1:19 V8 juice agar after incubation at 15 °C for 2–3 months. Asci are unconjugated and develop from the large “pulcherrima” cells that are commonly present and that assume a sphaeropedunculate shape (Fig. 4d). Asci measure 21–38 µm in length with the basal portion having a diameter of 4–8 µm. In contrast to many species of Metschnikowia, the basal portion of the ascus is somewhat elongated, rather than spherical (Fig. 4d). Asci are persistent and produce two closely paired acicular ascospores. Conjugation between mating types was tested on 1:19 V8 juice agar with incubation at 15 °C. Conjugants were usually detected within 2–3 days following mixing of complementary strains (Fig. 4c). The most active mating pair was NRRL Y-6148 (a) and NRRL Y-6066 (α).
Fermentation and growth reactions for this species are given in Table 2.
Type
NRRL Y-6064 is designated as the type strain of Metschnikowia rubicola. The strain is preserved as a metabolically inactive lyophilized preparation (the holotype) at the ARS Culture Collection, National Center for Agricultural Utilization Research, Peoria, IL, USA, and an isotype is preserved at the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands as CBS 15344. MycoBank number: MB 823165. Complementary mating types are NRRL Y-6148 (CBS 15347), mating type a, and NRRL Y-6066 (CBS 15340), mating type α. NRRL Y-6064, an ascosporogenous isolate, was isolated from a thimbleberry (Rubus parviflorus) growing in Sweet Home, Oregon, USA. Sources of other strains are given in Table 3.
Etymology
The species epithet rubicola refers to isolation of many of the strains from fruit of the thimbleberry, Rubus parviflorus, which was growing in Cape Perpetua and Sweet Home, OR, USA.
Ecology
Metschnikowia rubicola is presently known only from the state of Oregon, USA, and has been isolated primarily from thimbleberries and the flowers of red clover (Table 3). Presumably the species is insect disseminated.
Relatedness to other Metschnikowia species
On the basis of maximum likelihood analysis of concatenated sequences of D1/D2 and EF-1α, M. rubicola is most closely related to M. leonuri followed by M. shanxiensis, and M. chrysoperlae (Fig. 1). The relatedness between M. rubicola and M. leonuri will be discussed in the description of the species, which follows. M. rubicola differs from M. shanxiensis and M. chrysoperlae by substitutions in ITS, D1/D2, EF-1α (only for M. chrysoperlae) and RPB2 (Table 3). Although the number of substitutions with neighboring species is rather small, the 17 strains of M. rubicola do not differ from one another in the four sequences compared (Table 3). Notable is the sequence of the EF-1α gene, which for strains of M. rubicola contains 16–20 unresolved nucleotides (Table 3). Neighboring taxa M. leonuri, M. shanxiensis and M. chrosoperlae also have numerous unresolved positions in this gene sequence, which suggests the presence of two divergent copies. Of 20 unresolved nucleotide positions for NRRL Y-6064 and 18 for NRRL Y-6065, 18 are at the same location for each strain.
Fermentation and assimilation reactions of M. rubicola are so similar to other members of the M. pulcherrima clade that separation of species from phenotype is not certain.
Metschnikowia leonuri C. P. Kurtzman, C. J. Robnett & E. Basehoar sp. nov
Cell morphology
After 2 days on YM agar at 25 °C, cell morphology ranges from spherical (3–7 µm) to ovoidal (3–6 × 4–7 µm). Cell division is by multilateral budding and cells occur singly and in budded pairs (Fig. 5a). After 7 days at 25 °C, infrequent, poorly differentiated pseudohyphae formed under the coverglass on a Dalmau plate with corn meal agar (Fig. 5b), but true (septate) hyphae were not detected. Colony growth is low, white, semi-glistening, butyrous in texture and with irregular margins. Strains were not observed to form pulcherrimin on either YM agar or V8 juice agar.
Ascosporic state
Seven strains of M. leonuri are presently known and only one (NRRL Y-412) did not form ascospores (Table 4). This strain was tested with mating types of M. rubicola (NRRL Y-6066, NRRL Y-6148) but neither conjugations nor asci were observed in the mixtures after 2 months. For the other strains, ascosporulation was observed on 1:19 V8 juice agar after incubation at 15 °C for 1 week. Asci are unconjugated and develop from the large “pulcherrima” cells that are commonly present and assume a sphaeropedunculate shape (Fig. 5c). Asci measure 23–30 µm in length with the basal portion having a diameter of 6–8 µm. Asci are persistent and produce two closely paired acicular ascospores. It was not determined if the species is heterothallic.
Fermentation and growth reactions for this species are given in Table 2.
Type
NRRL Y-6546 is designated as the type strain of Metschnikowia leonuri. The strain is preserved as a metabolically inactive lyophilized preparation (the holotype) in the ARS Culture Collection, National Center for Agricultural Utilization Research, Peoria, IL, USA, and an isotype is preserved at the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands as CBS 15341. MycoBank number: MB 823166. NRRL Y-6546, an ascosporogenous isolate, was isolated from a motherwort plant (Leonurus cardiaca) growing in Peoria, Illinois, USA. Sources of other strains are given in Table 4.
Etymology
The species epithet leonuri refers to Leonurus cardiaca (mortherwort), the plant from which the type strain was isolated.
Ecology
Metschnikowia leonuri is presently known from flowers, a mushroom and the gummy exudate from a wild cherry tree, all from Midwestern USA (Table 4). If there is habitat specificity, flowers would seem to be the primary site.
Relatedness to other Metschnikowia species
When initially characterized from D1/D2 sequences, strains of M. leonuri were grouped with M. rubicola because they showed no nucleotide differences. However, small differences were noted in sequences for ITS, EF-1α and RPB2 (Table 4). These differences are not great, but they exceed differences ordinarily seen for strains of a species, suggesting that strains of M. leonuri should not be considered as divergent members of M. rubicola. The two species also show minor phenotypic differences, such as ascus shape and growth differences on cellobiose, l-sorbose, d-glucosamine and hexadecane. Strains of M. rubicola are known only from Oregon, whereas strains of M. leonuri are presently known only from Midwestern USA. Taken together, these small differences suggest the presence of two species.
As with M. rubicola, the EF-1α gene sequence for strains of M. leonuri shows 15–18 unresolved nucleotide positions, most of which are shared positions indicating the presence of two divergent copies of the EF-1α gene. In addition, the two available RPB2 sequences exhibit numerous unresolved nucleotides. NRRL Y-6463 has 55 unresolved positions, whereas NRRL Y-6464 has 63. Of these, 52 positions are shared. Consequently, it appears that M. leonuri may represent a hybrid species.
Metschnikowia anglica C. P. Kurtzman, C. J. Robnett & E. Basehoar sp. nov
Cell morphology
After 2 days on YM agar at 25 °C, cell morphology ranges from spherical (3–5 µm) to ellipsoidal (3–5 × 4–7 µm). Cell division is by multilateral budding and cells occur singly and in budded pairs (Fig. 6a). After 7 days at 25 °C, pseudohyphae and true (septate) hyphae were absent from under the coverglass on a Dalmau plate with corn meal agar. Colony growth is low, white, semi-glistening, butyrous in texture with a slightly irregular margin. Pulcherrimin was not observed on either YM agar or V8 juice agar.
Ascosporic state
A single strain is known for M. anglica and it forms ascospores, although sparingly. Ascosporulation was observed on 1:19 V8 juice agar after incubation at 15 °C for 11 weeks. Asci are unconjugated and develop from the large “pulcherrima” cells that are commonly present and assume a sphaeropedunculate shape (Fig. 6b). Asci measure 18–25 µm in length with the basal portion having a diameter of 5–6 µm. Asci are persistent and produce two closely paired acicular ascospores. It was not determined if the species is heterothallic.
Fermentation and growth reactions for this species are given in Table 2.
Type
NRRL Y-7298 is designated as the type strain of Metschnikowia anglica. The strain is preserved as a metabolically inactive lyophilized preparation (the holotype) in the ARS Culture Collection, National Center for Agricultural Utilization Research, Peoria, IL, USA, and an isotype is preserved at the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands as CBS 15342. MycoBank number: MB 823167. NRRL Y-7298 is an ascosporogenous isolate of the strain designated 489/1 Candida pulcherrima, which was received in February 1972 from Peter K. C. Austwick, Nuffield Institute of Comparative Medicine, Zoological Society of London, Regents Park, London, United Kingdom. The source of the isolate was not given.
Etymology
The species epithet anglica refers to England (UK), the apparent country of origin for the type strain.
Ecology
Unknown.
Relatedness to other Metschnikowia species
On the basis of maximum likelihood analysis of concatenated D1/D1 and EF-1α sequences, M. anglica is most closely related to Candida gelsemii (Fig. 1). The two species differ from one another by 44 substitutions and 30 indels in D1/D2 and 46 substitutions in EF-1α. Of the four gene sequences determined (ITS1-5.8S-ITS2, D1/D2, EF-1α, RPB2), unresolved nucleotides were found only in EF-1α, which exhibited two.
Because of similar fermentation and assimilation reactions, resolution of M. anglica from other species of Metschnikowia is uncertain when employing phenotype.
As required by the new code of nomenclature (International Code of Nomenclature for algae, fungi, and plants (Melbourne Code), McNeill et al. 2012), the following Candida species are transferred to Metschnikowia as new combinations because of their phylogenetic placement in that genus.
Metschnikowia chrysomelidarum (N.H. Nguyen, S.O. Suh, C.K. Erbil & M.Blackwell) C.P. Kurtzman, C.J. Robnett & E. Basehoar, f.a., comb. nov
Basionym: Candida chrysomelidarum N.H. Nguyen, S.O. Suh, C.K. Erbil & M. Blackwell (2006). Mycol. Res. 110: 352.
Type strain: NRRL Y-27749, CBS 9904.
MycoBank No.: MB 823223.
Metschnikowia gelsemii (M.A. Lachance) C.P. Kurtzman, C.J. Robnett & E. Basehoar, f.a., comb. nov
Basionym: Candida gelsemii M.A. Lachance (2007). Antonie van Leeuwenhoek 92: 40 (Manson et al. 2007).
Type strain: NRRL Y-48212, CBS 10509.
MycoBank No.: MB 823192.
Metschnikowia kofuensis (K. Mikata, K. Ueda-Nishimura, S. Goto, C.P. Kurtzman, M. Suzuki, D. Yarrow & T. Nakase) C.P. Kurtzman, C.J. Robnett & E. Basehoar, f.a., comb. nov
Basionym: Candida kofuensis K. Mikata, K. Ueda-Nishimura, S. Goto, C.P. Kurtzman, M. Suzuki, D. Yarrow & T. Nakase (1999). Microbiol. Cult. Coll. 15: 51.
Type strain: NRRL Y-27226, CBS 8058, IFO 10931.
MycoBank No.: MB 823195.
Metschnikowia picachoensis (S.O. Suh, C.M. Gibson & M. Blackwell) C.P. Kurtzman, C.J. Robnett & E. Basehoar, f.a., comb. nov
Basionym: Candida picachoensis S.O. Suh, C.M. Gibson & M. Blackwell (2004). Int. J. Syst.
Evol. Microbiol. 54: 1885.
Type strain: NRRL Y-27607, CBS 9804.
MycoBank No.: MB 823197.
Metschnikowia pimensis (S.O. Suh, C.M. Gibson & M. Blackwell) C.P. Kurtzman, C.J. Robnett & E. Basehoar, f.a., comb. nov
Basionym: Candida pimensis S.O. Suh, C.M. Gibson & M. Blackwell (2004). Int. J. Syst.
Evol. Microbiol. 54: 1887.
Type strain: NRRL Y-27619, CBS 9805.
MycoBank No.: MB 823205.
Metschnikowia rancensis (C. Ramírez & A. González) C.P. Kurtzman, C.J. Robnett & E. Basehoar, f.a., comb. nov
Basionym: Candida rancensis C. Ramírez & A. González (1984). Mycopathologia 88: 101.
Type strain: NRRL Y-48702, CBS 8174.
MycoBank No.: MB 823224.
Candida species proposed for transfer to the genus Clavispora as a new combination
For reference taxa, several species of the Clavispora clade were included in the phylogenetic analysis shown in Fig. 1. Besides Clavispora lusitaniae, type species of the genus, Cl. opuntiae is present along with Candida fructus and C. musae. These latter two species were proposed to be conspecific based on identical D1/D2 DNA sequences (Kurtzman and Robnett 1998), and Lu et al. (2004) found the two species to have identical ITS1-5.8S-ITS2 DNA sequences, thus further suggesting conspecificity. However, the two species were reported to differ by 14 substitutions and seven indels in the 18S rRNA gene sequence (Suzuki and Nakase 1999), indicating that the two taxa might not be conspecific. In the present study, DNA sequences for the two species were determined for 18S, EF-1α and RPB2 genes and repeated for D1/D2 and ITS. The only difference found was one substitution among 2148 nucleotides in the RPB2 gene sequence. The differences in the SSU sequences reported earlier were not detected. From these sequence comparisons, we conclude that C. fructus and C. musae are conspecific. GenBank accession numbers for C. fructus/C. musae sequences, respectively, are the following: D1/D2, U44810/U44814; ITS1-5.8S-ITS2, MG050897/MG050898; 18S rRNA, MG050883/MG050884; EF-1α, MG050963/MG050962; RPB2, MG050974/MG050975.
In a further comparison, type strains of C. fructus (NRRL Y-17072) and C. musae (NRRL Y-17088) were examined for ascospore formation following incubation alone and as a mixture on YM, 5% malt extract, YCBAS, RG and 1:19 V8 juice agar media at 15 and 25 °C with weekly microscopic inspections for 2 months. Neither conjugations nor ascospores were observed.
Both species were described in the same publication (Nakase 1971) and assigned to the genus Torulopsis, but later transferred to the genus Candida (Yarrow and Meyer 1978). Candida musae (Nakase) S.A. Meyer & Yarrow, 1978, Torulopsis fructus Nakase, 1971, and Torulopsis musae Nakase, 1971 (synonym) were listed as synonyms of Candida fructus (Offord and Kirk, 2017). In keeping with the requirements of the new code of nomenclature (International Code of Nomenclature for algae, fungi, and plants (Melbourne Code), McNeill et al. 2012), the following new combination is proposed for the name Candida fructus.
Clavispora fructus (T. Nakase) C.P. Kurtzman, C.J. Robnett & E. Basehoar, f.a., comb. nov
Basionym: Torulopsis fructus T. Nakase (1971). J. Gen. Appl. Microbiol. 17: 415.
Type strain: NRRL Y-17072, CBS 6380, IFO 1581.
MycoBank No.: MB 823206.
Synonyms:
Torulopsis fructus T. Nakase (1971).
Candida fructus (T. Nakase) S.A. Meyer & D. Yarrow (1978).
Torulopsis musae T. Nakase (1971).
Candida musae (T. Nakase) S.A. Meyer & D. Yarrow (1978).
Conclusions
Most yeast species are now routinely identified from unique sequences in the D1/D2 domains of the nuclear LSU rRNA gene and/or from divergence in ITS1-5.8S-ITS2 sequences. The preceding sequences have been viewed as reliable indicators of species identification, and strains of a species generally show no more than 0.5–1.0% sequence divergence (Kurtzman and Robnett 1998; Kurtzman et al. 2011; Vu et al. 2016). However, some exceptions have been noted. For example, Peterson and Kurtzman (1991) found no divergence between certain species of Saccharomyces, which later proved to be hybrids with each sharing the same gene for D1/D2, and Lachance et al. (2003) discovered strains of Clavispora lusitaniae that showed greater than expected D1/D2 divergence.
The findings of Sipiczki et al. (2013) are of particular interest to the current study. Sipiczki et al. (2013) observed that D1/D2 sequences from M. fructicola and M. andauensis contained a number of ambiguous nucleotides and wondered if these were the result of sequencing errors in the original studies (Kurtzman and Droby 2001; Molnár and Prillinger 2005) or resulted from other reasons. A repeat of the earlier work again gave a number of ambiguous nucleotides, which suggested that copies of divergent genes were present. Cloning revealed very few D1/D2 copies with identical sequences and the number of substitutions in different D1/D2 copies for M. andauensis ranged from 1 to 18, and substitutions in copies for M. fructicola ranged from 2 to 25. A neighbor-net analysis of cloned sequences showed that these species share a continuous pool of diverse repeats that appear to evolve by reticulate evolution. We observed ambiguous nucleotides in D1/D2 sequences for M. zizyphicola, M. pulcherrima and M. shanxiensis (Fig. 1). These three species along with M. fructicola and M. andauensis are members of the same subclade in Metschnikowia. ITS1-5.8-ITS2 sequences among strains of M. peoriensis show considerable variability, and based on preliminary data, it appears that multiple divergent copies of the ITS sequences are present, perhaps mimicking the results of the D1/D2 study of Sipiczki et al. (2013). Future work will need to include cloning of the ITS sequences to verify this premise.
In addition to the variation in ITS sequences found for M. peoriensis, we detected a substantial number of ambiguous nucleotides in the EF-1α sequence for M. rubicola, M. leonuri and other species of the M. pulcherrima clade as well as some nearby species (Fig. 1), which suggests the presence of two or more divergent copies of this gene. The presence of divergent copies of the RPB2 gene is also likely for strains of M. leonuri (Table 4).
The occurrence of hybrid species among the yeasts is poorly understood at this time, but studies of Saccharomyces indicate that some species show considerable introgression of genetic material from their neighbors (Hittinger et al. 2015; Dujon and Louis 2017). Consequently, from a taxonomic perspective, what degree of hybridization or non-introgressive genome chimerisation (Karanyicz et al. 2017) disqualifies a strain from being regarded as a distinct species? Clearly, there are no good answers at this time and thorough analysis of genome sequences will be required to provide guidance. Based on the apparent occurrence of multi-copy EF-1α genes in the M. pulcherrima clade, all species in this clade may be genomic chimeras. With the addition of M. rubicola and M. leonuri, there are now nine known species in the clade (Fig. 1), and this provides an opportunity to examine species boundaries in a clade well separated from the Saccharomyces clade, which has served as a model for studies in speciation, molecular genetics and evolution.
References
Banani H, Spadaro D, Zhang D, Matic S, Garibaldi A, Lodovica Gullino M (2015) Postharvest application of a novel chitinase cloned from Metschnikowia fructicola and overexpressed in Pichia pastoris to control brown rot of peaches. Int J Food Microbiol 199:54–61
Daniel HM, Lachance MA, Kurtzman CP (2014) On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie Van Leeuwenhoek 106:67–84
Dujon BA, Louis EJ (2017) Genome diversity and evolution in the budding yeasts (Saccharomycotina). Genetics 206:717–750
Guzmán B, Lachance MA, Herrera CM (2013) Phylogenetic analysis of the angiosperm-floricolous insect-yeast association: have yeast and angiosperm lineages co-diversified? Mol Phylogenet Evol 68:161–175
Hittinger CT, Rokas A, Bai FY, Boekhout T, Gonçalves P, Jeffries TW, Kominek J, Lachance MA, Libkind D, Rosa CA, Sampaio JP, Kurtzman CP (2015) Genomics and the making of yeast biodiversity. Curr Opin Genet Dev 35:100–109
Janisiewicz WJ, Tworkoski TJ, Kurtzman CP (2001) Biocontrol potential of Metschnikowia pulcherrima strains against blue mold of apple. Phytopathology 91:1098–1108
Kamienski T (1899) Notice préliminaire sur l’espèce de Metschnikowia (Monospora Metschn.). Trav Soc Imp Nat St Pétersbourg 30:363–364
Karanyicz E, Antunovics Z, Kallai Z, Sipiczki M (2017) Non-introgressive genome chimerisation by malsegregation in autodiploidised allotetraploids during meiosis of Saccharomyces kudriavzevii × Saccharomyces uvarum hybrids. Appl Microbiol Biotechnol 101:4617–4633
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kurtzman CP, Droby S (2001) Metschnikowia fructicola, a new ascosporic yeast with potential for biocontrol of postharvest fruit rots. Syst Appl Microbiol 24:395–399
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371
Kurtzman CP, Robnett CJ (2003) Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res 3:417–432
Kurtzman CP, Robnett CJ (2013) Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Res 13:23–33
Kurtzman CP, Fell JW, Boekhout T, Robert V (2011) Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier Science B.V, Amsterdam, pp 87–110
Lachance MA (2011) Metschnikowia Kamienski (1899). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier Science B.V, Amsterdam, pp 575–620
Lachance MA (2016) Metschnikowia: half tetrads, a regicide and the fountain of youth. Yeast 33:563–574
Lachance MA (2018) Personal reflections on Cletus P. Kurtzman (July 19 1938–November 27, 2017). Antonie Van Leeuwenhoek 111:1–4
Lachance MA, Daniel HM, Meyer W, Prasad GS, Gautam SP, Boundy-Mills K (2003) The D1/D2 domain of the large-subunit rDNA of the yeast species Clavispora lusitaniae is unusually polymorphic. FEMS Yeast Res 4:253–258
Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP (2011) Candida Berkhout. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier Science B.V, Amsterdam, pp 987–1277
Lu HZ, Jia JH, Wang QM, Bai FY (2004) Candida asparagi sp. nov., Candida diospyri sp. nov. and Candida qinlingensis sp. nov., novel anamorphic, ascomycetous yeast species. Int J Syst Evol Microbiol 54:1409–1414
Manson JS, Lachance MA, Thomson JD (2007) Candida gelsemii sp. nov., a yeast of the Metschnikowiaceae clade isolated from nectar of the poisonous Carolina jessamine. Antonie Van Leeuwenhoek 92:37–42
McNeill J, Barrie FR, Buck WR, Demoulin V, Greuter W, Hawkworth DL, Herendeen PS, Knapp S, Marhold K et al (2012) International code of nomenclature for algae, fungi, and plants (Melbourne Code). Regnum Veg, vol 154. Gantner Verlag, Koenigstein
Meena VS, Banoth L, Banerjee UC (2014) Demonstration of redox potential of Metschnikowia koreensis for stereoinversion of secondary alcohols/1,2-diols. Biomed Res Int 2014:410530. https://doi.org/10.1155/2014/410530 (Epub 2014 Jan 27)
Mikata K, Ueda-Nishimura K, Goto S, Kurtzman CP, Suzuki M, Yarrow D, Nakase T (1999) Reidentification of yeast strains deposited as Candida agrestis, with a description of Candida kofuensis sp. nov. Microbiol Cult Coll 15:49–57
Molnár O, Prillinger H (2005) Analysis of yeast isolates related to Metschnikowia pulcherrima using the partial sequences of the large subunit rDNA and the actin gene; description of Metschnikowia andauensis sp. nov. Syst Appl Microbiol 28:717–726
Nakase T (1971) New species of yeasts found in Japan. J Gen Appl Microbiol 17:409–419
Nguyen NH, Suh SO, Erbil CK, Blackwell M (2006) Metschnikowia noctiluminum sp. nov., Metschnikowia corniflorae sp. nov., and Candida chrysomelidarum sp. nov., isolated from green lacewings and beetles. Mycol Res 110:346–356
Offord LC, Kirk PM (2017) Saccharomycetes (version Jan 2016). In: Roskov Y, Abucay L, Orrell T, Nicolson D, Bailly N, Kirk PM, Bourgoin T, DeWalt RE, Decock W, De Wever A, van Nieukerken E, Zarucchi J, Penev L (eds) Species 2000 & ITIS catalogue of life, 28th November 2017. Digital resource at www.catalogueoflife.org/col. Species 2000: Naturalis, Leiden, the Netherlands. ISSN 2405-8858
Oro L, Ciani M, Comitini F (2014) Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J Appl Microbiol 116:1209–1217
Peterson SW, Kurtzman CP (1991) Ribosomal RNA sequence divergence among sibling species of yeasts. Syst Appl Microbiol 14:124–129
Piano S, Neyrotti V, Migheli Q, Gullino ML (1997) Biocontrol capability of Metschnikowia pulcherrima against Botrytis post-harvest rot of apple. Postharvest Biol Technol 11:131–140
Ramírez C, González A (1984) Two new amycelial Candida isolated from decayed wood in the evergreen rainy Valdivian forest of southern Chile. Mycopathologia 88:99–103
Sipiczki M (2006) Metschnikowia strains isolated from botrytized grapes antagonize fungal and bacterial growth by iron depletion. Appl Environ Microbiol 72:6716–6724
Sipiczki M, Pfliegler WP, Holb IJ (2013) Metschnikowia species share a pool of diverse rRNA genes differing in regions that determine hairpin-loop structures and evolve by reticulation. PLoS ONE 8(e67384):1–13
Sjamsuridzal W, Oetari A, Nakashima C, Kanti A, Saraswati R, Widyastuti Y, Ando K (2013) New species of the genus Metschnikowia isolated from flowers in Indonesia, Metschnikowia cibodasensis sp. nov. J Microbiol Biotechnol 23:905–912
Suh SO, Gibson CM, Blackwell M (2004) Metschnikowia chrysoperlae sp. nov., Candida picachoensis sp. nov. and Candida pimensis sp. nov., isolated from the green lacewings Chrysoperla comanche and Chrysoperla carnea (Neuroptera: Chrysopidae). Int J Syst Evol Microbiol 54:1883–1890
Suzuki M, Nakase T (1999) A phylogenetic study of ubiquinone Q8 species of the genera Candida, Pichia, and Citeromyces based on 18S ribosomal DNA sequence divergence. J Gen Appl Microbiol 45:239–246
Türkel S, Korukluoğlu M, Yavuz M (2014) Biocontrol activity of the local strain of Metschnikowia pulcherrima on different postharvest pathogens. Biotechnol Res Int 2014:397167. https://doi.org/10.1155/2014/397167
Vu D, Groenewald M, Szöke S, Cardinali G, Eberhardt U, Stielow B, deVries M, Verkley GJM, Crous PW, Boekhout T, Robert V (2016) DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud Mycol 85:91–105. https://doi.org/10.1016/j.simyco.2016.11.007 (Epub 2016 Nov 27)
Wang C, Liu Y, Zhang TT, Lu CG, Liu Y, Zhang DP, Liu WC (2017) Metschnikowia persici sp. nov., a novel protease-producing yeast species from China. Curr Microbiol 74:365–370
Wickerham LJ (1969) New homothallic taxa of Hansenula. Mycopathol Mycol Appl 37:15–32
Xue ML, Zhang LQ, Wang QM, Zhang JS, Bai FY (2006) Metschnikowia sinensis sp. nov., Metschnikowia zizyphicola sp. nov and Metschnikowia shanxiensis sp. nov., novel yeast species from jujube fruit. Int J Syst Evol Microbiol 56:2245–2250
Yarrow D, Meyer SA (1978) Proposal for amendment of the diagnosis of the genus Candida Berkhout nom. cons. Int J Syst Bacteriol 28:611–615
Acknowledgements
We thank Nathane Orwig for sequence determinations, and staff of the ARS Culture Collection for reference cultures. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by USDA. The USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Additional information
Cletus P. Kurtzman: Sadly, Clete Kurtzman died whilst this manuscript was under revision. An appreciation has been published (Lachance 2018).
Rights and permissions
About this article
Cite this article
Kurtzman, C.P., Robnett, C.J., Basehoar, E. et al. Four new species of Metschnikowia and the transfer of seven Candida species to Metschnikowia and Clavispora as new combinations. Antonie van Leeuwenhoek 111, 2017–2035 (2018). https://doi.org/10.1007/s10482-018-1095-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-1095-8