Abstract
DNA sequence analyses have demonstrated that species of the polyphyletic anamorphic ascomycete genus Candida may be members of described teleomorphic genera, members of the Candida tropicalis clade upon which the genus Candida is circumscribed, or members of isolated clades that represent undescribed genera. From phylogenetic analysis of gene sequences from nuclear large subunit rRNA, mitochondrial small subunit rRNA and cytochrome oxidase II, Candida auringiensis (NRRL Y-17674T, CBS 6913T), Candida salmanticensis (NRRL Y-17090T, CBS 5121T), and Candida tartarivorans (NRRL Y-27291T, CBS 7955T) were shown to be members of an isolated clade and are proposed for reclassification in the genus Groenewaldozyma gen. nov. (MycoBank MB 815817). Neighbouring taxa include species of the Wickerhamiella clade and Candida blankii.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Characterization of yeasts from DNA sequence analysis has resulted in accurate identification of species and an estimation of their phylogenetic placement (Kurtzman and Robnett 1998, 2013). DNA comparisons also showed that species of the genus Candida are distributed throughout the Saccharomycotina with some species seen as members of ascosporic genera, whereas others form isolated lineages that represent undescribed genera (Kurtzman and Robnett 1998). Because the D1/D2 LSU rRNA gene sequence used in many comparisons is relatively short (ca. 600 nucleotides), support for resulting clades is often weak, which has demonstrated the need for more extensive datasets to verify the placements suggested from D1/D2. In addition to the need for more robust datasets, until recently, reclassification of Candida species was hindered by provisions of the Botanical Code.
Classification of yeasts and other fungi has been governed by the rules of the International Code of Botanical Nomenclature (Vienna Code) (McNeill et al. 2006), which did not permit inclusion of teleomorphic and anamorphic species in the same genus, even if demonstrated to be congeneric. The latest edition of the Code, now entitled International Code of Nomenclature for algae, fungi, and plants (Melbourne Code) (McNeill et al. 2012), permits inclusion of anamorphic and teleomorphic species in the same genus, resulting in many new species combinations as anamorphs are transferred to teleomorphic genera. The genus Candida, with over 400 described species (Daniel et al. 2014) presents a unique problem. The genus was circumscribed on Candida vulgaris (= C. tropicalis), so the majority of Candida species are not members of this clade, but under the old Code, anamorphic ascomycete species were assigned to this genus because there were few other options. Now with changes in the Code and the application of phylogenetic analysis of gene sequences, it is possible to group related species, some of which are members of teleomorphic genera, some are members of Candida and some represent new genera. Where sufficient data are available, various Candida species have been transferred to new genera, such as Danielozyma, Deakozyma and Middelhovenomyces (Kurtzman and Robnett 2014). Multilocus DNA sequence analysis has demonstrated that Candida auringiensis, Candida salmanticensis and Candida tartarivorans are members of an isolated clade unrelated to other described genera, and it is proposed to assign these species to a new genus.
Materials and methods
Gene sequences for the nearly entire nuclear LSU rRNA, mitochondrial SSU rRNA and cytochrome oxidase II that were compared in this study had been determined earlier by Kurtzman and Robnett (2007) and that paper gives sequencing conditions, primers, and GenBank accession numbers for the sequences determined. For phylogenetic analysis, sequences were aligned using Muscle, which is in the MEGA version 5.2 software package (Tamura et al. 2011), and the alignments were visually adjusted. Phylogenetic relatedness among species was determined from the maximum likelihood program included in MEGA using the Hasegawa–Kishino–Yano model, and bootstrap support was determined from 1000 replicates. Species tested as the outgroup in the analyses included Trigonopsis variabilis, Tortispora caseinolytica and Schizosaccharomyces pombe.
Results and discussion
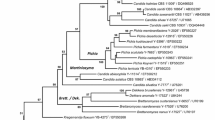
C. auringiensis and C. salmanticensis were recognized as an isolated pair of neighbouring species from analysis of gene sequences for D1/D2 domain LSU rRNA (Kurtzman and Robnett 1998) and for 18S rRNA (Suzuki et al. 1999). Soon after, Fonseca et al. (2000) added C. tartarivorans to the clade. A multilocus study by (Kurtzman and Robnett 2007) strongly supported the clade (100 % bootstrap) and provided information on neighbouring species. Now, due to adoption of the Melbourne Code, it is possible to transfer these three Candida species to a new genus (Fig. 1). which is described as follows.
Phylogenetic tree showing placement of the genus Groenewaldozyma among neighboring taxa. The tree was derived from maximum likelihood analysis of concatenated gene sequences from nearly entire nuclear LSU rRNA, mitochondrial small rRNA and cytochrome oxidase II. Bootstrap values >50 % are given at nodes and based on 1000 replicates. T. caseinolytica served as outgroup species in the analysis but replacement of T. caseinolytica with either T. variabilis or S. pombe had no effect on tree topology or branch support. All species are represented by type strains and are maintained in the ARS Culture Collection (NRRL), National Center for Agricultural Utilization Research, Peoria, IL USA
Groenewaldozyma Kurtzman gen. nov
Description of the genus
Growth is by multilateral budding on a narrow base and cells may be globose, ovoid or elongate. Pseudohyphae and true hyphae may be formed. An ascosporic state is unknown.
Species ferment glucose, galactose and trehalose, but fermentation of sucrose, maltose, lactose and raffinose is species and strain variable. Dien et al. (1996) reported some strains of C. auringiensis to ferment l-arabinose, a property useful for biomass utilization. Many of the hexoses, pentoses, disaccharides, sugar alcohols and organic acids commonly used in yeast taxonomy are assimilated, but there is no growth on methanol. Nitrate is not utilized as a sole source of nitrogen. Data for fermentation and growth reactions are from Lachance et al. (2011). The genus can be separated from other members of the Saccharomycotina by gene sequence analysis.
Phylogenetic placement: Saccharomycetales, Saccharomycotina, Ascomycota. The genus Groenewaldozyma, among currently recognized taxa, appears most closely related to species of the Wickerhamiella clade and to Candida blankii (Fig. 1, (Kurtzman and Robnett 2007).
Type species: G. auringiensis (Santa María) Kurtzman comb. nov.
MycoBank number: MB 815817.
Etymology: The genus is named in honor of Dr. Marizeth Groenewald, Centraalbureau voor Schimmelcultures Biodiversity Center, Utrecht, The Netherlands, for her many contributions to yeast systematics and yeast biodiversity.
Species of Candida proposed for transfer to the genus Groenewaldozyma as new combinations:
G. auringiensis (Santa María) Kurtzman comb. nov.
Basionym: C. auringiensis Santa María (1978). Commun Inst Nac Invest Agron, Serie General 3:55.
Type strain: NRRL Y-17674, CBS 6913.
Mycobank No.: MB 815818.
G. salmanticensis (Santa María) Kurtzman comb. nov.
Basionym: Torulopsis salmanticensis Santa María (1963). Antonie van Leeuwenhoek 29:330.
Type strain: NRRL Y-17090, CBS 5121.
Mycobank No.: MB 815819.
G. tartarivorans (Fonseca, Fell, Kurtzman & Spencer-Martins) Kurtzman comb. nov.
Basionym: C. tartarivorans (Fonseca et al. 2000) Int J Syst Evol Microbiol 50:390.
Type strain: NRRL Y-27291, CBS 7955.
Mycobank No.: MB 815820.
At present, Groenewaldozyma is a small genus with relatively little known about its ecology and geographical distribution. The description of G. auringiensis was based on three strains (CBS 6913T, CBS 6919, CBS 6920) that had been isolated from alpechin in Spain (Santa María 1978), but Lachance (Lachance et al. 2011) isolated additional apparently new species of Groenewaldozyma that are near G. auringiensis. These included UWO(PS)85–228.1.3 (GenBank AF530606) from sap fluxes of mesquite (Prosopis juliflora) growing in Arizona, USA and Baja California, Mexico, UWO(PS)93–324.2 from the rot of Agaves sp. in the Bahamas, and UWO(PS)99–304.7 (GenBank AF530605) from sap fluxes of the guapinol tree (Hymenacea courbaril) in Costa Rica. G. salmanticensis was also described by Santa María (1963) and had been isolated from alpechin in Spain. Since then, Lachance (Lachance et al. 2011) reported isolation of two additional strains, UWO(PS)85–301.1 from Drosophila carbonaria collected in sap flux of a mesquite (Prosopis juliflora), Arizona, USA, and UWO(PS)03–325y3 from sap flux of red oak (Quercus rubra), Ontario, Canada. The third species assigned to Groenewaldozyma, G. tartarivorans, was isolated from dried wine lees in Portugal (Fonseca et al. 2000). Three additional isolates are reported in GenBank: Y0 285 (GenBank LC015275) from fermentation silage of sugar cane, Brazil; IMUFRJ 51977 (GenBank FN428939) from a sugar cane field in Brazil; and NU28S61 (GenBank HM461710) from soil in Taiwan. From strain histories, it appears that distribution of Groenewaldozyma is worldwide. A majority of substrates from which strains have been isolated are characterized by low water activity suggesting additional species might be found in moderate to high osmotic habitats. Species of the neighbouring Wickerhamiella clade are often isolated from similar habitats (Lachance and Kurtzman 2011; Lachance et al. 2011) On the basis of D1/D2 analysis, there are presently 30 species assigned to the Wickerhamiella clade (Dayo-Owoyemi et al. 2014; Khunnamwong et al. 2014; Ren et al. 2014).
Phylogenetic placement of the genus Groenewaldozyma among other members of the Saccharomycotina is somewhat uncertain, but the multigene analysis of Kurtzman and Robnett (2007) placed the three species now assigned to Groenewaldozyma near Wickerhamiella and C. blankii. The analysis presented in Fig. 1 mirrors the earlier analysis, but with fewer included species. Consequently, Groenewaldozyma appears well isolated from Wickerhamiella and C. blankii, and it is likely that this separation will be maintained as more extensive datasets become available. The present work, as well as other recent studies (e.g., Kurtzman 2015; Kurtzman and Robnett 2014; Lachance and Kurtzman 2013), mark the beginning of the dissolution of the polyphyletic genus Candida into phylogenetically circumscribed genera, although placement of many Candida species must await more robust datasets.
References
Daniel HM, Lachance MA, Kurtzman CP (2014) On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie van Leeuwenhoek 106:67–84
Dayo-Owoyemi I, Rosa CA, Rodrigues A, Pagnocca FC (2014) Wickerhamiella kiyanii f.a., sp. nov. and Wickerhamiella fructicola f.a., sp. nov., two yeasts isolated from native plants of atlantic rainforest in Brazil. Int J Syst Evol Microbiol 64:2152–2158
Dien BS, Kurtzman CP, Saha BC, Bothast RJ (1996) Screening of l-arabinose fermenting yeasts. Appl Biochem Biotechnol 57(58):233–242
Fonseca Á, Fell JW, Kurtzman CP, Spencer-Martins I (2000) Candida tartarivorans sp. nov., an anamorphic ascomycetous yeast with the capacity to degrade l(+)-and meso-tartaric acid. Int J Syst Evol Microbiol 50:389–394
Khunnamwong P, Surussawadee J, Jindamorakot S, Limtong S (2014) Wickerhamiella siamensis f.a., sp. nov., an endophytic and epiphytic yeast species isolated from sugar cane leaf. Int J Syst Evol Microbiol 64:3849–3855
Kurtzman CP (2015) Description of Martiniozyma gen. nov. and transfer of seven Candida species to Saturnispora as new combinations. Antonie van Leeuwenhoek 108:803–809
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73:331–371
Kurtzman CP, Robnett CJ (2007) Multigene phylogenetic analysis of the Trichomonascus, Wickerhamiella and Zygoascus yeast clades, and the proposal of Sugiyamaella gen. nov. and fourteen new species combinations. FEMS Yeast Res 7:141–151
Kurtzman CP, Robnett CJ (2013) Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Res 13:23–33
Kurtzman CP, Robnett CJ (2014) Three new anascosporic genera of the Saccharomycotina: Danielozyma gen. nov., Deakozyma gen. nov. and Middelhovenomyces gen. nov. Antonie van Leeuwenhoek 105:933–942
Lachance MA, Kurtzman CP (2011) Wickerhamiella van der Walt. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier Science B.V., Amsterdam, pp 891–897
Lachance MA, Kurtzman CP (2013) The yeast genus Tortispora gen. nov., description of Tortispora ganteri sp. nov., Tortispora mauiana f.a., sp. nov., Tortispora agaves f.a., sp. nov., Tortispora sangerardii f.a., sp. nov., Tortispora cuajiniquilana f.a., sp. nov., Tortispora starmeri f.a., sp. nov., and Tortispora phaffii f.a., sp. nov., reassignment of Candida caseinolytica to Tortispora caseinolytica f.a., comb. nov., emendation of Botryozyma, and assignment of Botryozyma, Tortispora gen. nov., and Trigonopsis to the family Trigonopsidaceae fam. nov. Int J Syst Evol Microbiol 63:3104–3114
Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP (2011) Candida Berkhout. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier Science B.V., Amsterdam, pp 987–1277
McNeill J, Barrie FR, Burdet HM, Demoulin V, Hawksworth DL, Marhold K, Nicolson DH, Prado J, Silva PC, Skog JE, Wiersema JH, Turland NJ (2006) International Code of Botanical Nomenclature (Vienna Code) Regnum Veg, 146. Gantner, Ruggell, Liechtenstein
McNeill J, Barrie FR, Buck WR, Demoulin V, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Marhold K et al (2012) International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). Regnum Veg, 154. Koelz Scientific Books, Koenigstein
Ren YC, Wang Y, Chen L, Ke T, Hui FL (2014) Wickerhamiella allomyrinae f.a., sp. nov., a yeast species isolated from the gut of the rhinoceros beetle Allomyrina dichotoma. Int J Syst Evol Microbiol 64:3856–3861
Santa María J (1963) New melibiose-utilizing yeasts isolated from “alpechin”. Antonie van Leeuwenhoek 29:329–343
Santa María J (1978) Biotaxonomic studies on yeast. Commun Inst Nac Invest Agron, Serie General 3:1–61
Suzuki M, Suh SO, Sugita T, Nakase T (1999) A phylogenetic study on galactose-containing Candida species based on 18S ribosomal DNA sequences. J Gen Appl Microbiol 45:229–238
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Acknowledgments
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kurtzman, C.P. Description of Groenewaldozyma gen. nov. for placement of Candida auringiensis, Candida salmanticensis and Candida tartarivorans . Antonie van Leeuwenhoek 109, 1041–1045 (2016). https://doi.org/10.1007/s10482-016-0703-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0703-8