Abstract
An ovoid to rod-shaped, phototrophic, purple non-sulfur bacterium was isolated from a sediment sample of a hot spring in Tibet, China. Cells of strain YIM 73036T were Gram-stain negative, non-motile and multiplied by binary fission. Strain YIM 73036T grew optimally at pH 7.0–7.5 at 37–45 °C. Growth occurred in 0.5–3.5% (w/v) NaCl. Vitamins were not required for growth. The presence of photosynthesis genes pufL and pufM were shown and photosynthesis pigments were formed. Bacteriochlorophyll α, the bacteriopheophytin and carotenoids were present as photosynthetic pigments. Internal cytoplasmic membranes were of the lamellar type. The organism YIM 73036T was able to grow chemo-organoheterophically, chemo-lithoautotrophically and photo-organoheterotrophically but photo-lithoautotrophic and fermentative growth were not demonstrated. Phylogenetic analysis on the basis of 16S rRNA gene sequences showed that strain YIM 73036T is closely related to Rhodobacter blasticus ATCC 33485T (96.65% sequence similarity) and clustered with species of the genus Rhodobacter of the family Rhodobacteraceae. Whole-genome sequence analyses based on the average nucleotide BLAST identity (ANI < 82%) indicated that this isolate belongs to a novel species. The genomic DNA G+C content of organism YIM 73036T was determined to be 66.0 mol%. Strain YIM 73036T contained Q-10 as the predominant ubiquinone and C18:1ω7c, C18:1ω7c 11-methyl and C18:0 as the major fatty acids. The major polar lipids were phosphatidylglycerol, diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine and unidentified phospholipid. Differential phenotypic and chemotaxonomic properties, together with the phylogenetic distinctiveness, demonstrated that strain YIM 73036T is distinguishable from other species of the genus Rhodobacter. On the basis of the data presented, strain YIM 73036T is considered to represent a novel species of the genus Rhodobacter, for which the name Rhodobacter thermarum sp. nov. [type strain YIM 73036T (= KCTC 52712T = CCTCC AB 2016298T)] is proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Rhodobacter belongs to the family Rhodobacteraceae of the class Alphaproteobacteria in the phylum Proteobacteria. Members of this genus are Gram-stain negative and contain bacteriochlorophyll-a, as the core of photosynthesis reaction system RC complexes in phototrophic bacteria (Zhang et al. 2016); phosphatidylglycerol (PG), phosphatidylethanolamine (PE) and phosphatidylcholine (PC) are the major polar lipids of species of the genus Rhodobacter (Raj et al. 2013). At the time of writing, the genus Rhodobacter comprises 15 recognized species names: Rhodobacter aestuarii, Rhodobacter azotoformans, Rhodobacter blasticus, Rhodobacter capsulatus, Rhodobacter johrii, Rhodobacter maris, Rhodobacter megalophilus, Rhodobacter ovatus, Rhodobacter sphaeroides, Rhodobacter veldkampii, Rhodobacter vinaykumarii, Rhodobacter viridis, Rhodobacter sediminis and the recently described Rhodobacter lacus and Rhodobacter azollae (Subhash and Lee 2016; Suresh et al. 2017). Species of this genus have been isolated from different environmental samples, such as a brown-coloured microbial mat (Venkata et al. 2009), semi-arid tropical soils (Girija et al. 2010), marine habitats (Venkata et al. 2008; Srinivas et al. 2007), polluted pond sediment (Srinivas et al. 2008), mud of a stream (Raj et al. 2013) and lagoon sediments (Subhash and Lee 2016) and pond (Suresh et al. 2017). The family Rhodobacteraceae members, particularly the genus Rhodobacter is a very heterogeneous assemblage of phototrophic bacteria with a large number of interspersing chemotrophic bacteria and their evolutionary relationships are not well established. Based on 16S rRNA gene sequence phylogenetic analysis, Rhodobacter species are grouped into five monophyletic clusters, each of which comprise one to seven species. In this study, we describe isolation and characterization of a non-motile, Gram-stain negative phototrophic Alphaproteobacterium, designated YIM 73036T, which was isolated from a sediment sample collected at Qucai Geothermal Field, in Tibet hot spring, China. Comparative 16S rRNA gene sequence analysis indicated that this strain is closely related to members of the genus Rhodobacter. The aim of the present work was to determine the exact taxonomic position of strain YIM 73036T by using a polyphasic characterization that included determination of chemotaxonomic and other phenotypic properties and phylogenetic investigation based on 16S rRNA gene sequences and at the genomic level.
Materials and methods
Isolation and preservation
The sediment sample was collected from Qucai Geothermal Field (pH 7.0, temperature 65.7 °C, 30°39′58.5″N, 91°35′28.8″E) in August 2014. For the isolation, 3 g of sediment sample was taken into a flask with 30 ml sterile water. The flask was kept incubated in a shaker (37 °C, 200 r.p.m., 2 h). The resultant suspension was diluted to 10−2 dilution, and 0.5 ml aliquots of the diluted suspension was spread on R2A agar (pH 7.0–7.2) plates with the following composition (0.6 g peptone, 0.6 g yeast extract, 0.6 g glucose, 0.6 g casamino acids, 0.6 g soluble starch, 0.3 g sodium pyruvate, 0.3 g K2HPO4·7H2O, 0.005 g MgSO4·7H2O, 15.0 g agar per litre). The isolation plates were incubated at 45 °C for 1 week. The strain YIM 73036T was maintained as pure cultures on R2A agar medium at 45 °C, and stored as a glycerol suspension (20%, w/v) at − 80 °C. R. blasticus ATCC 33485T and Tabrizicola aquatica RCRI19T were grown under the same conditions and used as reference strains for comparative taxonomic work.
Phenotypic characterization
Morphological characteristics of strain YIM 73036T were determined by transmission electron microscopy (JEM-2100; JEOL) after the culture was grown on R2A agar at 45 °C for 2 days. The internal membrane structures were determined from exponentially growing cultures by using a transmission electron microscope (JEM-2100; JEOL) after the cells had been processed (Hanada et al. 2002). Gram staining was carried out by using the standard Gram’s reaction and was confirmed by a non-staining procedure (Gregersen 1978). Growth was tested at 20, 25, 37, 45, 50 and 55 °C in R2A broth medium. Tolerance of salt was tested by supplementing R2A broth medium with various concentrations of NaCl ranging from 0.5 to 5% (w/v) at intervals of 0.5%. The pH response was determined in R2A broth medium adjusted between pH 4.0 and 11.0 (with interval of 1.0 pH unit) using buffer system as described by Xu et al. (2005). Nitrogen source utilization tests were carried out as described by Gordon et al. (1974). The ability to utilize various carbon sources, production of acid from sugars and physiological profile of the strain YIM 73036T were studied using API ZYM, API 20 NE and API 50 CH B/L kits according to the manufacturer’s (BioMérieux) instructions.
To test phototrophic growth, the bacterium was incubated in Pfennig medium (Pfennig and Truper 1992), supplemented with sodium pyruvate (0.3%, w/v) and NH4Cl (0.12%) as carbon and nitrogen sources, respectively, under light exposure (2400 l×) and anaerobic conditions at 30 ± 2 °C. Photo-lithoautotrophic growth was investigated under anaerobic condition and light exposure (2400 l×) with Na2S (0.5 mM), Na2S2O3 (0.5 mM) and NaHCO3 (0.1%, w/v). The bacterium was incubated in aerobic dark conditions in Pfennig medium in the presence of sodium pyruvate (0.3%, w/v) as the only carbon source to determinate chemo-organoheterotrophic growth. After that, the growth was investigated under aerobic and dark conditions with Na2S2O3 (0.5 mM) and NaHCO3 (0.1%, w/v) also under anaerobic, dark conditions with pyruvate (0.3%, w/v) in order to determine chemo-lithoautotrophic and fermentative growth, respectively. Vitamin (biotin, niacin, p-aminobenzoic acid, thiamine and vitamin B12) requirement was tested by replacing yeast extract with single and also combinations of vitamins as growth factors. Determination of oxidase activity was carried out using 1% (w/v) tetramethyl-p-phenylenediamine as described by Kovacs (1956). Catalase activity was tested using 3% (w/v) H2O2 by assessing bubble production as the positive result. Other biochemical characteristics including hydrolysis of aesculin, casein, chitin, gelatin and Tweens (20, 40, 60 and 80), H2S production and nitrate reduction were observed as previously described (MacFaddin 1976; Gonzalez et al. 1978; Smibert and Krieg 1994).
Chemotaxonomy
Biomass used for chemical studies was obtained from cultures grown on R2A agar plates for 3 days at 45 °C. Polar lipids were extracted, separated by two-dimensional TLC and identified using previously described procedures (Minnikin et al. 1979; Collins and Jones 1980). For the cellular fatty acid analysis, strain YIM 73036T and related type strains were harvested from growth on R2A [Difco (pH 7.0)] plates at 37 °C for 2 days. The cellular fatty acids were extracted, methylated and analyzed by using the Microbial Identification System (Sherlock Version 6.1; MIDI database: TSBA6) (Sasser 1990). Quinones were isolated as described by Collins et al. (1977) and separated by HPLC (Kroppenstedt 1982). To extract carotenoids, 100 ml of T5 medium in semi-aerobically condition cultures were centrifuged at 9000 rpm for 10 min at 4 °C. After separation of supernatant, the pellet was mixed with acetone-methanol (7:3 v/v) solution containing 0.1% butylhydroxytoluene as antioxidant. The pelleted cells were then frozen in liquid nitrogen and thawed in room temperature to improve the extraction yield. This step was repeated several times and finally was followed by centrifugation at 12,000×g for 15 min at 4 °C. The solvent was evaporated under a stream of nitrogen gas and the pigments were dissolved in 10 ml of acetone (containing 0.1% BHT). The identification of carotenoids were performed by comparing retention time, UV–VIS spectra and characteristics of the mass spectra (protonated molecule ([M+H]+). All of the carotenoids were monitored at 450 nm with a UV–visible detector (Naziri et al. 2014). Xcalibur 2.0 SR2 software (copyright Thermo Electron Corporation 1998–2006) was used for data analysis. In order to confirm the presence of bacteriochlorophyll a and pigments, UV–VIS absorption spectra of the strain was measured with a spectrophotometer (Shimadzu UV-1800 Series, Kyoto, Japan). Moreover, the carotenoid composition comparison, was determined by C18-HPLC analysis among strains YIM 73036T, R. blasticus ATCC 33485T and T. aquatica RCRI19T under similar phototrophical conditions (Ramana et al. 2010).
The genomic DNA G+C contents were determined by HPLC after enzymatic degradation (Mesbah et al. 1989) using E. coli strain DH5α as the reference.
Molecular characterization
Extraction of genomic DNA and PCR amplification of the 16S rRNA gene sequences were performed as previously described (Cui et al. 2001; Li et al. 2007). The resulting 16S rRNA gene sequence was compared with available 16S rRNA gene sequences of cultured species from GenBank via the BLAST program and from the EzBioCloud server databases (Yoon et al. 2017a, b). The genetic ability to form a photosynthetic apparatus was tested by the presence of pufL and pufM genes. PCR amplification of the pufL and pufM genes was performed using the primer set used previously with the forward primer (67F) 5′-TTC GAC TTY TGG RTN GGN CC-3′ and the reverse primer (781R) 5′-CCA KSG TCC AGC GCC AGA ANA-3′ (Tank et al. 2009). Phylogenetic trees were generated using three tree-making algorithms, neighbour-joining (Saitou and Nei 1987), maximum-likelihood (Felsenstein 1981) and maximum parsimony (Fitch 1971) methods in the MEGA version 5.0 software package (Tamura et al. 2011). Kimura’s two-parameter model (Kimura 1980) was used to calculate evolutionary distance matrices of the neighbour-joining method and maximum-likelihood method. The topology of the phylogenetic trees was evaluated by the bootstrap resampling method of Felsenstein (1985) with 1000 resamplings. Average Nucleotide Identity (ANI) was calculated between the draft genome sequence of strain YIM 73036T and closely related reference type strains by using online software (Yoon et al. 2017a, b).
Genome sequencing, assembly and annotation
Draft genome sequencing of strain YIM 73036T was performed on a Illumina HiSeq 2000 platform (Illumina, USA) at MajorBio Shang Hai, China. Genome assembly of the raw sequence data generated was performed with SOAPdenovo2 (Luo et al. 2012).
Nucleotide sequence accession numbers
The Whole Genome Shotgun project of strain YIM 73036T has been deposited at DDBJ/EMBL/GenBank under the accession QMJY00000000. Genomes for the reference strains were downloaded from NCBI.
Results and discussion
Phenotypic characteristics
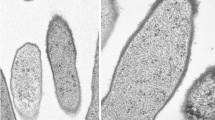
The colonies of strain YIM 73036T were small with a convex surface. The bacterium produces glassy-colored colonies on marine agar medium with 0.5% NaCl at 30 °C. The colonies change to pink over time. Strain YIM 73036T was found to be Gram-stain negative. The isolate YIM 73036T was able to grow on T5, R2A and marine agar media. Cells of the organism YIM 73036T were rod shaped with 0.7–1.1 × 1.8–3.4 µm (supplementary Fig. S1). The strain showed growth at 20–55 °C, optimum growth at temperature 37–45 °C. Growth for strain YIM 73036T was observed at pH 6.0–8.5 (optimum, 7.0–7.5). The novel isolate was positive for oxidase and catalase activities. Strain YIM 73036T was positive for hydrolysis of Tween 20, but negative for milk coagulation, hydrolysis of chitin, gelatin and Tweens (40, 60, 80), and also H2S production tests. The bacterium was able to grow chemo-organoheterophically [aerobic, dark, in the presence of pyruvate (0.3% w/v)], chemo-lithoautotrophically [aerobic, dark, Na2S2O3 (0.5 mM) and NaHCO3 (0.1% w/v)] (very weak) and photo-organoheterotrophic growth [anaerobic conditions, in the light 2400 l×, pyruvate (0.3% w/v)] but photo-lithoautotrophic growth [anaerobic, light 2400 l×, Na2S (0.5 mM), Na2S2O3 (0.5 mM) and NaHCO3 (0.1%, w/v)] and fermentative growth [anaerobic, dark, pyruvate (0.3%, w/v)] do not occur.
During photosynthetic growth (Imhoff et al. 1984), the colour of the cell suspension of both strains YIM 73036T and R. blasticus ATCC 33485T were yellow–brown. The whole-cell extract absorption spectrum (Fig. S4) of strain YIM 73036T showed maximam absorptions at 375, 480, 681, 759 and 846 nm, confirming the presence of bacteriochlorophyll a and pigments (Girija et al. 2010). According to the LC–MS data at 450 nm, the major caretenoid of strain YIM 73036T in the semi-aerobically growth is spheroidenone (Chi et al. 2015). Also HPLC analysis determined that strains YIM 73036T and the closely related species R. blasticus ATCC 33485T showed the same pattern for the major carotenoids (Fig.S5) implying the presence of spheroidene and sphoroidenone as major carotenoids (Girija et al. 2010). Interestingly we didn’t observe any peak for pigments in T. aquatica RCRI19T in this condition. Lamellar type of internal cytoplasmic membranes were present (Fig S1).
In the API ZYM system, the isolate displayed positive results for activities of alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, naphthol-AS-BI-phosphohydrolase, β-galactosidase, α-glucosidase and β-glucosidase. Applying API 20NE kit β-galactosidase and β-glucosidase were positive but negative results were obtained for indole production, fermentation (glucose), arginine dihydrolase, urease, and hydrolysis of gelatin. The reduction of nitrate was negative. Arabinose, glucose, mannose, manitol, maltose and adipic acid are assimilated but N-acetyl-glucosamine, capric acid, potassium gluconat, malate, trisodium citrate, and phenylacetic acid are not assimilated. According to API 50CH kit results, the bacterium can produce acid from d-glucose, d-fructose, d-sorbitol, l-arabinose, salicine, d-saccharose (sucrose), d-lyxose, d-xylose, d-mannitol, esculin, d-fucose, methyl-α-d-monno pyranoside, d-lactose (bovine origin), d-trehalose, d-melezitose, and d-cellobiose, but not from glycerol, d-arabinose, erythritol, d-tagatose, l-lyxose, d-adonitol, d-mannose, l-sorbose, l-rhamnose, dulcitol, inositol, methyl-α-d-mannapyanoside, l-rhamnose, dulcitol, inositol, N-acetylglucosamine, amygdalin, arbutin, d-celiobiose, d-maltose, d-melibiose, inulin, d-raffinose, amidon (starch), glycogen, xylitol, gentiobiose, d-turanose, l-fucose, l-arabitol, potassium gluconate, potassium 2-ketogluconate, d-ribose, methyl-β-d-xylopyranoside, d-galactose, methyl-α-d-glucopyranoside, potassium 5-keto gluconate, and d-arabitol. Selective differentiating characteristics with closely related type strains and the genus Rhodobacter species are listed in Tables 1 and S3.
Chemotaxonomical characteristics
The polar lipid profile consists of PG, diphosphatidylglycerol (DPG), PE, PC, two unidentified phospholipids and two unidentified aminophospholipids (Fig. S3). The cellular fatty acids (> 1% of total fatty acids) of strain YIM 73036T consisted of C18:1ω7c (50.9%), C18:1ω7c 11-methyl (19.6%), C18:0 (10.8%), C16:0 (7.4%), C18:0 3–OH (4.9%) and C10:0 3–OH (2.5%) (Table S1). The predominant cellular fatty acids particularly C18:1ω7c and C18:1ω7c 11-methyl were also reported in other members of the genus Rhodobacter. In the present study, the fatty acid (C18:1ω7c) was predominant in the novel isolate, Rhodobacter thermarum YIM 73036T and R. blasticus CGMCC 1.3365T which was consistent with the previous study, in Rhodobacter sediminis N1T, R. capsulatus KACC 15298T and Rhodobacter viridis KCTC 15167T (Subhash and Lee 2016). Subhash and Lee (2016) also reported the dominant fatty acids (C16:1ω6c/C16:1ω7c) in R. sediminis N1T, R. capsulatus KACC 15298T and R. viridis KCTC 15167T which were not detected in R. thermarum YIM 73036T and R. blasticus CGMCC 1.3365T in the present study. Also in the present study, C18:1ω7c 11-methyl was a major component in R. thermarum YIM 73036T, in a small amount in R. blasticus CGMCC 1.3365T, while in the earlier study, not detected or in trace amount in R. sediminis N1T, R. capsulatus KACC 15298T and R. viridis KCTC 15167T (Subhash and Lee 2016).
Detailed fatty acid profiles of strain YIM 73036T and the two closely related reference type strains are given in Table S1. The major respiratory lipoquinone was ubiquinone-10 (Q-10), which is consistent with those reported for the genus Rhodobacter. The DNA G+C contents of YIM 73036T were determined to be 66.0 mol%.
Molecular characteristics
Based on 16S ribosomal gene sequence and comparison to 16S rRNA genes in the EzTaxon-e server, strain YIM 73036T is closely related to R. blasticus ATCC 33485T (96.65%) and T. aquatica RCRI19T (95.85%). The ANI values between the draft genome of strain YIM 73036T and the reference type strains were found to be below 95% (Supplementary Table S2), showed that the isolate YIM 73036T belongs to a novel species. The phylogenetic trees generated using methods based on neighbour-joining, maximum-parsimony and maximum-likelihood algorithms showed that strain YIM 73036T formed a distinct phylogenetic lineage within the genus Rhodobacter (Fig. 1 and supplementary Figs. S6 and S7 respectively).
Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences showing the relationships of strain YIM 73036T with related members of the family Rhodobacteraceae. Bootstrap percentages based on 1000 resamplings are listed at the nodes. Only bootstrap values above 50% are shown at branch points. Asterisks indicate branches that were also recovered in the maximum-parsimony and maximum likelihood trees. 16S rRNA gene sequence of Rhodospirillum rubrum ATCC 11170T (NR_074249) was used as an outgroup. Bar, 0.02 substitutions per nucleotide position
The morphological features, chemotaxonomic properties and 16S rRNA gene sequence similarity profiles clearly indicated that strain YIM 73036T is a member of the genus Rhodobacter. Strain YIM 73036T exhibited several physiological and chemotaxonomic characteristics of the genus Rhodobacter: containing bacteriochlorophyll-a and PG, PE and PC which are the major polar lipids of species of the genus Rhodobacter (Raj et al. 2013). Characteristics that differentiate the strain (YIM 73036T) from related type strains of the genus Rhodobacter include differences in growth conditions (temperature and pH ranges for growth), utilization of carbon and nitrogen sources, as well as the proportions of some fatty acids (Tables 1 and S1). The fatty acid profiles of strain YIM 73036T and related reference type strains were similar, but there were differences in the proportions of some fatty acids. In particular, strain YIM 73036T was characterized by having a considerable amount of C18:1ω7c11-methyl (19.6%) and C18:0 (10.8%) which were minor components (2.0%) and (3.5%), respectively, in closely related reference strain R. blasticus CGMCC 1.3365T (Supplementary Table S1). The polar lipid profile of YIM 73036T was similar to that of R. blasticus CGMCC 1.3365T in that PG, DPG, PE, PC and unidentified phospholipid were the major polar lipids, but it could be distinguished from the reference strain R. blasticus CGMCC 1.3365T by the nature and proportion of the differences between the other polar lipids (Fig. S3). In particular, two unidentified aminophospholipids were detected in strain YIM 73036T, which were absent from the closely related reference type strain R. blasticus CGMCC 1.3365T. Based on these properties, strain YIM 73036T merit classification as a novel species of the genus Rhodobacter, for which the name Rhodobacter thermarum sp. nov. is proposed.
Description of Rhodobacter thermarum sp. nov.
Rhodobacter thermarum (ther.ma’rum. L. gen. pl. n. thermarum of hot springs).
Cells are Gram-stain negative and ovoid to rod-shaped (0.7–1.1 × 1.8–3.4 μm in size) and multiply by binary fission. The colonies are small with a convex surface. The bacterium produces glassy-colored colonies on marine agar medium with 0.5% NaCl at 30 °C. The colonies change to pink over time. Growth occurs at 20–55 °C, pH 6.0–8.0 and in the presence of up to 3.5% (w/v) NaCl. Positive for catalase and oxidase activities. Hydrolyzes Tween 20, but not casein, starch, or Tweens 40, 60 and 80. Bacteriochlorophyll-a, bacteriopheophytin and spheroidene and spheroidenone are the photosynthetic pigments. Internal cytoplasmic membranes are of the lamellar type. The predominant respiratory quinone is ubiquinone Q-10 and the G+C content of the genomic DNA of the type strain is 66.0 mol%. The major cellular fatty acids (> 10%) are C18:1ω7c,C18:1ω 7c 11-methyl and C18:0. The polar lipids consisted of phosphatidylglycerol, diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine, unidentified phospholipids and aminophospholipids.
The type strain YIM 73036T (= KCTC 52712T = CCTCC AB 2016298T) was isolated from a sediment sample collected from a hot spring in western Tibet, China. The GenBank accession number of the 16S rRNA gene sequence of strain YIM 73036T is KY608089, and its draft genome sequence accession number is QMJY00000000. The taxon number of the strain in the digital protologue is TA00508.
References
Chi SC, Mothersole DJ, Dilbeck P, Niedzwiedzki DM, Zhang H, Qian P, Vasilev C, Grayson KJ, Jackson PJ, Martin EC, Li Y, Holten D, Neil Hunter C (2015) Assembly of functional photosystem omplexes in Rhodobacter sphaeroides incorporating carotenoids from the spirilloxanthin pathway. Biochim Biophys Acta 1847:189–201
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2, 4-diaminobutyric acid. J Appl Bacteriol 48:459–470
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Cui XL, Mao PH, Zeng M, Li WJ, Zhang LP, Xu LH, Jiang CL (2001) Streptimonospora salina gen. nov., sp. nov., a new member of the family Nocardiopsaceae. Int J Syst Evol Microbiol 51:357–363
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Girija KR, Sasikala C, Ramana CV, Spröer C, Takaichi S, Thiel V, Imhoff JF (2010) Rhodobacter johrii sp. nov., an endospore producing cryptic species isolated from semi-arid tropical soils. Int J Syst Evol Microbiol 60:2099–2107
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. an amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715
Gordon RE, Barnett DA, Handerhan JE, Pang CH-N (1974) Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol 24:54–63
Gregersen T (1978) Rapid method for distinction of Gram-negative from Gram-positive bacteria. Eur J Appl Microbiol Biotechnol 5:123–127
Hanada S, Takaichi S, Matsuura K, Nakamura K (2002) Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int J Syst Evol Microbiol 52:187–193
Imhoff JF, Trüper HG, Pfennig N (1984) Rearrangement of the species and genera of the phototrophic “purple nonsulfur bacteria”. Int J Syst Evol Microbiol 34(3):340–343
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kovacs N (1956) Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 178:703
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J Liq Chromatogr 5:2359–2367
Li WJ, Xu P, Schuman P, Zhang YQ, Pukall R, Xu LH, Stackebrandt E, Jiang CL (2007) Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China) and emended description of the genus Georgenia. Int J Syst Evol Microbiol 57:1424–1428
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Xhi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18
Macfaddin JF (1976) Biochemical tests for identification of medical bacteria. Williams & Wilkins Co, Philadelphia
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin D, Collins M, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95
Naziri D, Hamidi M, Hassanzadeh S, Tarhriz V, Zanjani BM, Nazemyieh H, Hejazi MA, Hejazi MS (2014) Analysis of carotenoid production by Halorubrum sp. TBZ126; an extremely halophilic archeon from Urmia Lake. Adv Pharm Bull 4(1):61
Pfennig N, Truper HG (1992) The family Chromatiaceae. In: Balows A, Tru¨per HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes, 2nd edn. Springer, New York, pp 3200–3221
Raj PS, Ramaprasad EV, Vaseef S, Sasikala C, Ramana CV (2013) Rhodobacter viridis sp. nov., a phototrophic bacterium isolated from mud of a stream. Int J Syst Evol Microbiol 63:181–186
Ramana VV, Sasikala Ch, Ramaprasad EVV, Ramana ChV (2010) Description of Ectothiorhodospira salini sp. nov. J Gen Appl Microbiol 56:313–319
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20:16
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. Am Soc Microbiol, Washington, pp 607–654
Srinivas TN, Kumar PA, Sasikala C, Ramana CV, Imhoff JF (2007) Rhodobacter vinayakumarii sp. nov., a marine phototrophic alphaproteobacterium from tidal waters, and emended description of the genus Rhodobacter. Int J Syst Evol Microbiol 57:1984–1987
Srinivas TNR, Kumar PA, Sasikala C, Spröer C, Ramana CV (2008) Rhodobacter ovatus sp. nov., a phototrophic alphaproteobacterium isolated from a polluted pond. Int J Syst Evol Microbiol 58:1379–1383
Subhash Y, Lee SS (2016) Rhodobacter sediminis sp. nov., isolated from lagoon sediments. Int J Syst Evol Microbiol 66:2965–2970
Suresh G, Sailaja B, Ashif A, Dave Bharti P, Sasikala Ch, Ramana Ch (2017) Description of Rhodobacter azollae sp. nov. and Rhodobacter lacus sp. nov. Int J Syst Evol Microbiol 67:3289–3295
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tank M, Thiel V, Imhoff JF (2009) Phylogenetic relationship of phototrophic purple sulfur bacteria according to pufL and pufM genes. Int Microbiol 12:175–185
Tarhriz V, Thiel V, Nematzadeh G, Hejazi MA, Imhoff JF, Hejazi MS (2013) Tabrizicola aquatica gen. nov. sp. nov., a novel alphaproteobacterium isolated from Qurugo¨l Lake nearby Tabriz city, Iran. Antonie Van Leeuwenhoek 104:1205–1215
Venkata RV, Sasikala C, Ramana C (2008) Rhodobacter maris sp. nov., a phototrophic alphaproteobacterium isolated from amarine habitat of India. Int J Syst Evol Microbiol 58:1719–1722
Venkata RV, Anil KP, Srinivas TN, Sasikala C, Ramana C (2009) Rhodobacter aestuarii sp. nov., a phototrophic alphaproteobacterium isolated from an estuarine environment. Int J Syst Evol Microbiol 59:1133–1136
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH, Xu H, Jiang CL (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family Oxalobacteraceae isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017a) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Yoon SH, Ha SM, Lim JM, Kwon SJ, Chun J (2017b) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Zhang H, Harrington LB, Lu Y, Prado M, Saer R, Rempel D, Blankenship RB, Gross ML (2016) Native mass spectrometry characterizes the photosynthetic reaction center complex from the purple bacterium Rhodobacter sphaeroides. J Am Soc Mass Spectrom 28:87–95
Acknowledgements
We are grateful to Professor Yu-Guang Zhou (CGMCC, China) and Professor Jung-Sook Lee (KCTC, Korea) for their kindly providing the reference type strains. This work was supported by the Key Project of International Cooperation of Ministry of Science and Technology (MOST, China) (No. 2013DFA31980), Science and Technology Infrastructure work project (No. 2015FY110100), National Natural Science Foundation of China (No. 31470139) and Basic Scientific Research Service Fee Project in Colleges and Universities (No. 17lgjc19). W-J Li was also supported by Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (2014). We also acknowledge Molecular Medicine Research Center, Biomedicine Institute, Tabriz University of Medical Sciences (Tabriz, Iran) for its support.
Author information
Authors and Affiliations
Contributions
IUK, NH, MX and WJL conducted this study. IUK, NH, MSH, VT and MX performed the experiments. XYZ and WJL supervised the experiments. IUK, NH, MML, and WDX wrote the manuscript. All of the authors assisted in writing the manuscript, discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no direct or indirect conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Khan, I.U., Habib, N., Xiao, M. et al. Rhodobacter thermarum sp. nov., a novel phototrophic bacterium isolated from sediment of a hot spring. Antonie van Leeuwenhoek 112, 867–875 (2019). https://doi.org/10.1007/s10482-018-01219-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-01219-7