Abstract
A Gram-positive, non-motile, asporogenous and aerobic bacterium, designated YIM 98012T, was isolated from a salt lake in China. Strain YIM 98012T was found to be catalase and oxidase positive. Optimal growth of strain YIM 98012T was observed at 37 °C and pH 7.0 and it was found to grow in the presence of 5–20% (w/v) NaCl (optimum 10% NaCl). Phylogenetic analysis based on the 16S rRNA gene sequence indicated that the novel strain is affiliated with the family Bacillaceae of the phylum Firmicutes and that it shares high (94.7%) sequence similarity with Alteribacillus persepolensis DSM 21632T and does not show sequence similarities of more than 94.0% to known members of other related genera. The major fatty acids (> 10%) were identified as anteiso-C15:0, anteiso-C17:0, iso-C16:0 and C16:0. The genomic DNA G+C content was determined to be 41.0 mol% and the dominated respiratory quinone was identified as MK-7. The cell wall peptidoglycan of strain YIM 98012T was found to contain meso-diaminopimelic acid, while the polar lipids profile was found to include diphosphatidylglycerol, phosphatidylglycerol and phosphatidylcholine. Based on physiological and chemotaxonomic characteristics, strain YIM 98012T is concluded to be the type strain of the type species of a novel genus in the family Bacillaceae for which the name Aidingibacillus halophilus gen. nov., sp. nov. is proposed. The type strain is YIM 98012T (= KCTC 33868T = DSM 104332T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The family Bacillaceae in the order Bacillales is a large taxonomic group currently containing more than 55 genera with many different physiological features (http://www.bacterio.net/). Based on 16S rRNA gene sequence and chemotaxonomic analyses, many bacteria which were originally placed in the genus Bacillus have been reclassified as members of novel genera or transferred to other genera (Claus and Berkeley 1986). Halophilic microorganisms are also often found in the family Bacillaceae, such as members of the genera Halobacillus (Spring et al. 1996), Oceanobacillus (Lu et al. 2001), Ornithinibacillus (Mayr et al. 2006), Terribacillus (An et al. 2007), Sediminibacillus (Carrasco et al. 2008), Streptohalobacillus (Wang et al. 2011), Saliterribacillus (Amoozegar et al. 2013), Aquibacillus (Amoozegar et al. 2014) and Sinibacillus (Yang and Zhou 2014).
In this study, we describe a novel moderately halophilic, Gram-positive, asporogenous bacterial strain, designated YIM 98012T, isolated from sediment from Ayding Lake, a saline lake, in Xinjiang, China. The comparative analysis of 16S rRNA gene sequences showed that the strain phylogenetically belongs to the family Bacillaceae but does not fall into any currently defined genera. Strain YIM 98012T was subjected to a polyphasic taxonomic characterisation based on physiological and chemotaxonomic characteristics and identified as the type strain of the type species of a novel genus, Aidingibacillus gen. nov.
Materials and methods
Isolation of the bacterial strain and culture conditions
Strain YIM 98012T was isolated from sediment samples collected from Ayding Lake, which is situated in the southern part of Turpan Basin in Xinjiang Uygur Autonomous Region, north-west China. The lake is 155 m below sea level, and is the lowest place in China and the second lowest place in the world, next only to the Dead Sea (− 391 m) of Jordan. The pH of the saline soil ranges between 8.0 and 8.5 and the salinity ranges from 36 to 112 (g kg−1 soil). The major ion contents of the sediment samples were measured (g kg−1 soil): Na+ (10.6–34.3), Cl− (16.2–82.0), SO4 2− (6.8–23.5), K+ (0.06–0.1), Ca2+ (2.2–4.5), Mg2+ (0.2–0.8) and HCO3 − (0.1–0.4). The novel strain was isolated by plating on the Cellulose–casein multi-salts medium described by Tang et al. (2008) and incubating at 37 °C aerobically. The strain was maintained on GTY medium slants containing 10% NaCl (w/v) at 4 °C and as 20% (w/v) glycerol suspensions at − 20 °C. The GTY medium contained [(l distilled water)−1]: 0.5 g tryptone, 2 g yeast extract, 1 g glucose, 0.5 g CaCO3, 100 g NaCl and 15 g agar. The pH of the medium was adjusted to pH 7.5 (Tang et al. 2009).
Morphological, physiological, biochemical and chemotaxonomic characterisation
For phenotypic characterisation of strain YIM 98012T, standard tests were performed. The recommended minimal standards for describing new taxa of aerobic, endospore-forming bacteria were followed (Logan et al. 2009). Cell morphology was examined with a microscope equipped with phase-contrast optics (Olympus) and transmission electron microscopy (JEM2100; JEOL), using cells from exponentially growing cultures. Gram staining was carried out by the standard Gram reaction and was confirmed using the KOH lysis test method (Cerny 1978). Gliding motility was tested in 0.2% GTY medium by using a hanging-drop technique (Bernardet et al. 2002). Motility was confirmed by puncture inoculation in semisolid agar medium. Spore formation was tested by staining with malachite green.
The temperature range (0, 4, 10, 15, 20, 28, 30, 37, 40, 45, 50 and 55 °C) and pH range (4.0–10.0, at intervals of 1 pH unit) for colony growth were determined by incubating the isolate for 2 weeks on GTY medium containing 10% NaCl. The buffers for pH tests were prepared according to the method described previously by Tang et al. (2009). The salt concentration (NaCl) range from 0 to 30% (w/v) at intervals of 1% were tested by using GTY agar without adding any salts as the basal medium. Catalase activity was determined by production of bubbles after the addition of a drop of 3% (v/v) H2O2. Oxidase activity was observed by oxidation of tetramethyl-p-phenylenediamine. Reduction of nitrate, the methyl red and Voges–Proskauer tests, hydrolysis of aesculin, gelatin, casein, starch and Tweens 20, 40, 60 and 80 were determined as described by Dong and Cai (2001). Enzyme activities and acid production from carbohydrates were determined by API 20NE, API ZYM and API 50CH strips (bioMérieux) according to the instructions of the manufacturer. Strains were prepared using pre-warmed sterile saline medium (10% NaCl), within the density range specified by the manufacturer. Carbon-source utilisation tests were performed according to the methods described by Shirling and Gottlieb (1966). Nitrogen-source utilisation tests were analysed as described by Williams et al. (1983). Antibiotic sensitivity was explored by placing commercial antibiotic discs (OXOID) on GTY agar plates that had been spread with the isolates and then incubated at 37 °C for 7 days. Anaerobic growth was tested for up to 2 weeks on GTY medium containing 10% NaCl in a jar containing AnaeroPack-Anaero (Mitsubishi Gas Chemical Co, Inc), which works as an O2 absorber and CO2 generator.
Diaminopimelic acid isomers were analysed using the method of Komagata and Suzuki (1988). For the analysis of fatty acids, strains YIM 98012T was cultured on tryptic soy agar (TSA; Difco) containing 10% NaCl at 37 °C for 72 h and fatty acid methyl esters were extracted and gas chromatography was performed as described by Sasser (1990) using the microbial identification system (MIDI) according to standard protocols. Isoprenoid quinones were extracted and purified as described by Collins et al. (1977). The purified menaquinones were dissolved in acetone and separated by reverse-phase HPLC. Polar lipids were extracted and identified by two-dimensional TLC (Minnikin and Goodfellow 1979).
Phylogenetic analysis and DNA G+C content analysis
Genomic DNA was prepared by using a commercial DNA extraction kit (Quick-DNA™ Bacterial Miniprep Kit; Zymo research). The G+C contents of the DNAs were determined by reverse-phase HPLC (Mesbah et al. 1989). An approximately 1500 bp long fragment of the 16S rRNA gene was amplified from the extracted DNA by using bacterial universal primers specific to the 16S rRNA gene: 27F and 1492R (Escherichia coli numbering system, Weisburg et al. 1991). The obtained sequence was compared with 16S rRNA reference gene sequences retrieved from the GenBank and EMBL database by BLAST search and similarity searches were performed using the EzBioCloud Database (http://eztaxon-e.ezbiocloud.net/) (Yoon et al. 2016); phylogenetically closely related sequences belonging to the family Bacillaceae were downloaded from the NCBI database. These sequences were aligned using the ClustalW program integrated in the MEGA 6.0 software (Tamura et al. 2013). Alignment gaps and ambiguous bases were not taken into consideration when 1524 bases of the 16S rRNA gene were compared. Evolutionary distances (distance options according to Kimura’s two-parameter model) were calculated. Phylogenetic analyses were performed using three tree-making algorithms that were the neighbour-joining, maximum-likelihood and maximum-parsimony methods. A phylogenetic tree was constructed using the neighbour-joining method using MEGA version 6.0. The topology of the phylogenetic tree was evaluated by the bootstrap resampling method with 1000 replicates (Felsenstein 1985).
Results and discussion
Morphological, physiological and biochemical characteristics
Cells of strain YIM 98012T grown on GTY medium containing 10% NaCl were observed to be straight rods with dimensions 0.4–1.0 μm in width and 2.4–5.2 μm in length, and devoid of flagella (Supplementary Fig. S1). Spores were not observed following growth under a range of conditions. Gliding motility was not observed by light microscopy. The strain was determined to be positive for catalase, oxidase and hydrolysis of Tween 80 but negative for Voges–Proskauer and methyl red tests, casein, urea and starch hydrolysis, and hydrolysis of Tweens 20, 40, 60. In the API 20NE system, nitrate was not reduced to nitrite, gelatin was not liquefied, H2S and indole are not produced. Arginine dihydrolase, lysine dihydrolase, lysine decarboxylase, β-galactosidase (PNPG) and tryptophan deaminase activities were not detected. Other physiological properties are given in the species description. The following substrates were found to be utilised as sole carbon and nitrogen sources: amygdalin, l-arabinose, citrate, cellobiose, d-fructose, d-galactose, glycerol, d-lactose, d-mannitol, d-mannose, d-maltose, l-rhamnose, d-ribose, d-sorbitol, l-salicin, strach, d-trehalose, d-xylose, l-asparagine, l-cysteine, glycine, l-glutamic acid, l-histidine, l-leucine, methionine, l-proline, l-tyrosine and l-valine, but not d-melibiose, d-raffinose, d-saccharose, xylitol, l-alanine or l-phenylalanine. In the API ZYM system, strain YIM 98012T was found to be positive for esterase (C4), esterase lipase (C8) and naphthol-AS-BI-phosphohydrolase-β-glucosidase and negative for alkaline phosphatase lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, β-galactosidase, α-galactosidase, α-glucosidase β-glucosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase. Acid was found to be produced from d-fructose, aesculin citrate defer and d-raffinose (API 50 CHB). Strain YIM 98012T was found to be sensitive to ampicillin (10 μg), amphozone (30 mg), azithromycin (15 μg), bacitracin (10 U), clarithromycin(15 μg), cefalotin (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), erythromycin (15 μg), gentamicin (10 μg), kanamycin (30 μg), lincomycin (2 μg), novobiocin (5 μg), neomycin (30 μg), polymyxin B (300 U), penicillin G (10 U), netilmicin (10 μg), roxithromycin (15 μg), streptomycin (10 μg), compound sulphonamides (300 μg), tobramycin (10 μg), tetracycline (30 μg), vancomycin (30 μg), but resistant to amikacin (30 μg). The detailed physiological and biochemical characteristics of strain YIM 98012T and of closely related type strains, including of two type strains of the related genus Alteribacillus, Bacillus salarius BH169T and Salibacterium halotolerans KCTC 33658T are listed in Table 1. Strain YIM 98012T can be distinguished from Alteribacillus bidgolensis P4BT and Alteribacillus persepolensis HS136T by absence of spore formation, salinity range for growth and acid production. Strain YIM 98012T differs from the B. salarius BH169T by hydrolysis of aesculin and casein. Strain YIM 98012T can be distinguished from S. halotolerans KCTC 33658T by its negative Voges–Proskauer test reaction and hydrolysis Tween 80 and its acid production of d-fructose.
Phylogenetic analysis
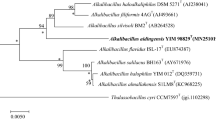
The nearly complete sequence (1524 bp, GenBank Accession Number KY427816) of the 16S rRNA gene was obtained and this sequence was subjected to phylogenetic analysis. According to the EzBioCloud Database, strain YIM 98012T has high 16S rRNA gene sequence similarity to members of the family Bacillaceae and is closely related to the members of the genus Alteribacillus. Strain YIM 98012T shares high sequence similarity with A. persepolensis HS136T (94.7%), A. bidgolensis P4BT (94.2%), followed by Bacillus piscicola NR1-3-2T (93.4%), B. salarius BH169T (93.0%). Strain YIM 98012T showed 16S rRNA gene sequence similarity of less than 93.0% with all other current members of the family Bacillaceae with validly published names (Fig. 1). In the neighbour-joining, maximum-likelihood and maximum-parsimony phylogenetic trees based on 16S rRNA gene sequences, strain YIM 98012T forms a well separated branch, among the members of the genera Alteribacillus, Salibacterium, Bacillus, Salipaludibacillus and Scopulibacillus, which belong to the family Bacillaceae. Phylogenetic analysis using the neighbour-joining method showed that the isolate formed a distinct phylogenetic line, with an 83% bootstrap value, from the genera Alteribacillus, Salibacterium and Bacillus (Fig. 1). The phylogenetic position was confirmed from trees generated using the maximum-likelihood algorithms with high bootstrap values (51%). Phylogenetic trees based on maximum-parsimony algorithms was also constructed, and although they demonstrated a different tree topology, the relationships among the members of the family Bacillaceae remained similar (Supplementary Fig. S2). Yarza et al. (2014) proposed a threshold of 94.5% sequence identity for delineation of a new genus based on 16S rRNA gene analyses. Thus, strain YIM 98012T can be considered to represent a novel genus in the family Bacillaceae.
Neighbor-joining phylogenetic tree showing the relationships between strain YIM 98012T and representatives of the family Bacillaceae. Filled circles indicate that the corresponding nodes were also found in the tree generated with maximum-likelihood algorithm. Bootstrap percentages (based on 1000 replications) > 50% are shown at branching points. Bar, 0.01 substitutions per nucleotide position. The 16S rRNA gene sequence of Paenibacillus polymyxa IAM 13419T(AB042063) was arbitrarily chosen as outgroup
Chemotaxonomic characteristics
The predominant cellular fatty acids (> 10%) of strain YIM 98012T were identified anteiso-C15:0 (41.6%), anteiso-C17:0 (21.0%), iso-C16:0 (10.9%) and C16:0 (10.8%), which is similar to those found in other members of the family Bacillaceae. Anteiso-C15:0 is usually predominant in the type strains belonging to the family Bacillaceae. Marginal quantitative differences were observed in the profile of the fatty acids compared with the type strains of A. bidgolensis P4BT, A. persepolensis HS136T, B. salarius BH169T and S. halotolerans KCTC 33658T (Table 1). However, iso-C15:0 which is a major fatty acid in type strains of phylogenetically closely related Alteribacillus species, was observed to be a minor fatty acid in strain YIM 98012T (Table 2).
The major respiratory quinone of YIM 98012T was identified as menaquinone-7 (MK-7) with minor amounts of menaquinone-6 (MK-6), which in accordance with the characteristics of closely related genera. The polar lipids of strain YIM 98012T were determined to be composed of diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), phosphatidylcholine (PC), a glycolipid (GL) and five unidentified phospholipids (PL) (Supplementary Fig. S3). The presence of DPG, PC and the presence of unidentified phospholipids in the strain YIM 98012T lipid profile helps distinguish the strain from the type strains of the genera Alteribacillus (Didari et al. 2012; Amoozegar et al. 2009). The presence of PC, PL and GL and the absence of unidentified aminolipids in strain YIM 98012T can differentiate it from the members of the genus Salibacterium (Reddy et al. 2015). The presence of PC is a discriminative characteristic compared with the type species of closely related genera (Table 1). This characteristic supports the conclusion that strain YIM 98012T represents a novel genus.
Strain YIM 98012T was found to possess meso-diaminopimelic acid in the cell wall peptidoglycan. The DNA G+C content of strain YIM 98012T was determined to be 41.0 mol%, which is in the range of the five phylogenetically related species, but lower than that of Salibacterium halotolerans (48.4 mol%).
Based on this polyphasic analysis, from the distinct phylogenetic position and combination of genotypic and phenotypic characteristics, strain YIM 98012T cannot be assigned to any previously recognised bacterial genus and thus can be described as representing a novel species within a new genus, here named Aidingibacillus halophilus gen. nov., sp. nov. Characteristics that distinguish strain YIM 98012T from the members of related genera within family Bacillaceae are shown in Table 1. The Digital Protologue database (Rosselló-Móra et al. 2017) TaxoNumber for strain YIM 98012T is GA00042.
Description of Aidingibacillus gen. nov.
Aidingibacillus (Ay.ding.i.ba.cil’lus. N.L. n. Ayding, a lake, located in Xinjiang province of north-west China; L. masc. n. bacillus a small rod; N.L. masc. n. Aidingibacillus a rod-shaped microbe isolated from Ayding lake).
A member of the family Bacillaceae, phylum Firmicutes, according to 16S rRNA gene sequence analyses. Cells are rod shaped, Gram stain positive and strictly aerobic. Endospores are not formed. Moderately halophilic, growing over a wide range of NaCl concentrations with optimal growth in the presence of 10% (w/v) NaCl. Catalase and oxidase positive. The major respiratory quinone is menaquinone 7 (MK-7). The predominant cellular fatty acids are anteiso-C15:0 and anteiso-C17:0. The peptidoglycan contains meso-diaminopimelic acid as the diagnostic diamino acid. The polar lipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylcholine, a glycolipid and five unidentified phospholipids. The DNA G+C content of the type strain of the type species is 40.1 mol%. The type species is Aidingibacillus halophilus.
Description of Aidingibacillus halophilus sp. nov.
Aidingibacillus halophilus (ha.lo’phi.lus. Gr. n. hals, halos salt; Gr. adj. philos loving; N.L. masc. adj. halophilus, salt loving).
In addition to the characteristics given in the genus description, colonies are cream, flat and opaque, with slightly irregular edges on GTY medium. Cells are straight rods with dimensions 0.4–1.0 × 2.4–5.2 μm. Growth occurs at 18–50 °C, pH 4.0–10.0 and 5–20% (w/v) NaCl, with the optimal growth at 37 °C, pH 7.0 and 10% NaCl. Nitrate is not reduced. Urease test is negative. Voges–Proskauer and methyl red tests are negative. Indole and H2S are not produced. Tweens 20, Tweens 40, Tweens 60, gelatin, casein or starch are not hydrolysed, while hydrolysis of Tween 80 is positive. The major fatty acids are anteiso-C15:0, anteiso-C17:0, iso-C16:0, C16:0, iso-C15:0, iso-C17:0 and iso-C14:0.
The type strain is YIM 98012T (= KCTC 33868T = DSM 104332T), isolated from Ayding Lake, a salt lake, in Xinjiang, China.
References
Amoozegar MA, Sánchez-Porro C, Rohban R, Hajighasemi M, Ventosa A (2009) Bacillus persepolensis sp. nov., a moderately halophilic bacterium from a hypersaline lake. Int J Syst Evol Microbiol 59:2352–2358
Amoozegar MA, Bagheri M, Didari M, Fazeli SAS, Schumann P, Sánchez-Porro C, Ventosa A (2013) Saliterribacillus persicus gen. nov., sp. nov., a moderately halophilic bacterium isolated from a hypersaline lake. Int J Syst Evol Microbiol 63:345–351
Amoozegar MA, Bagheri M, Didari M, Mehrshad M, Schumann P, Spröer C, Sánchez-Porro C, Ventosa A (2014) Aquibacillus halophilus gen. nov., sp. nov., a moderately halophilic bacterium from a hypersaline lake, and reclassification of Virgibacillus koreensis as Aquibacillus koreensis comb. nov. and Virgibacillus albus as Aquibacillus albus comb. nov. Int J Syst Evol Microbiol 64:3616–3623
An SY, Asahara M, Goto K, Kasai H, Yokota A (2007) Terribacillus saccharophilus gen. nov., sp. nov. and Terribacillus halophilus sp. nov., spore-forming bacteria isolated from field soil in Japan. Int J Syst Evol Microbiol 57:51–55
Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Carrasco IJ, Márquez MC, Xue Y, Ma Y, Cowan DA, Jones BE, Grant WD, Ventosa A (2008) Sediminibacillus halophilus gen. nov., sp. nov., a moderately halophilic, Gram-positive bacterium from a hypersaline lake. Int J Syst Evol Microbiol 58:1961–1967
Cerny G (1978) Studies on aminopeptidase for the distinction of Gram-negative from Gram-positive bacteria. Eur J Appl Microbiol Biotechnol 5:113–122
Claus D, Berkeley RCW (1986) Genus Bacillus Cohn 1872 174AL. In: Sneath PHA, Mair NS, Sharpe ME, Holt JG (eds) Bergey’s manual of systematic, vol 2. Williams & Wilkins, Baltimore, pp 1105–1139
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Didari M, Amoozegar MA, Bagheri M, Schumann P, Sproer C, Sanchez-Porro C, Ventosa A (2012) Alteribacillus bidgolensis gen. nov., sp. nov., a moderately halophilic bacterium from a hypersaline lake, and reclassification of Bacillus persepolensis as Alteribacillus persepolensis comb. nov. Int J Syst Evol Microbiol 62:2691–2697
Dong X, Cai M (2001) Manual of systematic and determinative bacteriology. Academic Press, Beijin (in chinese)
Felsenstein J (1985) Conference limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Komagata K, Suzuki KI (1988) 4 Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Lim JM, Jeon CO, Lee SM, Lee JC, Xu LH, Jiang CL, Kim CJ (2006) Bacillus salarius sp. nov., a halophilic, spore-forming bacterium isolated from a salt lake in China. Int J Syst Evol Microbiol 56:373–377
Logan NA, Berge O, Bishop AH, Busse HJ, De Vos P, Fritze D, Heyndrickx M, Kampfer P, Rabinovitch L, Salkinoja-Salonen MS, Seldin L, Ventosa A (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59:2114–2121
Lu J, Nogi Y, Takami H (2001) Oceanobacillus iheyensis gen. nov., sp. nov., a deep-sea extremely halotolerant and alkaliphilic species isolated from a depth of 1050 m on the Iheya Ridge. FEMS Microbiol Lett 205:291–297
Mayr R, Busse HJ, Worliczek H, Ehling-Schulz M, Scherer S (2006) Ornithinibacillus gen. nov., with the species Ornithinibacillus bavariensis sp. nov. and Ornithinibacillus californiensis sp. nov. Int J Syst Evol Microbiol 56:1383–1389
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95
Reddy SV, Thirumala M, Sasikala C, Ramana CV (2015) Salibacterium halotolerans gen. nov., sp. nov., a bacterium isolated from a salt pan, reclassification of Bacillus qingdaonensis as Salibacterium qingdaonense comb. nov. and Bacillus halochares as Salibacterium halochares comb. nov. Int J Syst Evol Microbiol 65:4270–4275
Rosselló-Móra R, Trujillo ME, Sutcliffe IC (2017) Introducing a digital protologue: a timely move towards a database-driven systematics of archaea and bacteria. Antonie Van Leeuwenhoek 110:455–456
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI Inc, Newark
Shirling E, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Spring S, Ludwig W, Marquez M, Ventosa A, Schleifer KH (1996) Halobacillus gen. nov., with descriptions of Halobacillus litoralis sp. nov. and Halobacillus trueperi sp. nov., and Transfer of Sporosarcina halophila to Halobacillus halophilus comb. nov. Int J Syst Evol Microbiol 46:492–496
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tang SK, Tian X, Zhi XY, Cai M, Wu JY, Yang LL, Xu LH, Li WJ (2008) Haloactinospora alba gen. nov., sp. nov., a halophilic filamentous actinomycete of the family Nocardiopsaceae. Int J Syst Evol Microbiol 58:2075–2080
Tang SK, Zhi XY, Wang Y, Wu JY, Lee JC, Kim CJ, Lou K, Xu LH, Li WJ (2009) Haloactinobacterium album gen. nov., sp. nov., a halophilic actinobacterium, and proposal of Ruaniaceae fam. nov. Int J Syst Evol Microbiol 60:2113–2119
Wang X, Xue Y, Ma Y (2011) Streptohalobacillus salinus gen. nov., sp. nov., a moderately halophilic, Gram-positive, facultative anaerobe isolated from subsurface saline soil. Int J Syst Evol Microbiol 61:1127–1132
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Williams ST, Goodfellow M, Alderson G, Wellington EM, Sneath PH, Sackin MJ (1983) Numerical classification of Streptomyces and related genera. J Gen Microbiol 129:1743–1813
Yang G, Zhou S (2014) Sinibacillus soli gen. nov., sp. nov., a moderately thermotolerant member of the family Bacillaceae. Int J Syst Evol Microbiol 64:1647–1653
Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzéby J, Amann R, RossellóMöra R (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2016) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 31400438, 31270055, 31560026).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Y., Chunyu, WX., Jiang, GQ. et al. Aidingibacillus halophilus gen. nov., sp. nov., a novel member of the family Bacillaceae . Antonie van Leeuwenhoek 111, 601–608 (2018). https://doi.org/10.1007/s10482-017-0980-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0980-x