Abstract
A Gram-positive, rod-shaped, strictly aerobic bacterium, strain WSY08-1T, was isolated from a salt mine in Wensu county, Xinjiang province, China. Spherical to ellipsoidal endospores were observed to be formed in terminal swollen sporangia. Strain WSY08-1T was found to be able to grow at 20–45 °C (optimum 37 °C), 0–10 % (w/v) NaCl (optimum 4 %, w/v) and pH 6.0–9.0 (optimum 7.0). Catalase and oxidase activities were observed to be positive. The cell-wall peptidoglycan of strain WSY08-1T was found to contain meso-diaminopimelic acid. Menaquinone-7 (MK-7) was identified as the predominant isoprenoid quinone. The polar lipids were found to consist of phosphatidylglycerol, diphosphatidylglycerol, an unknown glycolipid, two unknown phospholipids and an unknown lipid. The major cellular fatty acids were identified as anteiso-C15:0 and anteiso-C17:0. The DNA G+C content was determined to be 36.9 mol%. Analysis of the 16S rRNA gene sequence showed that strain WSY08-1T is closely related to Aquibacillus halophilus B6BT, Aquibacillus koreensis BH30097T and Aquibacillus albus YIM 93642T (97.6, 96.9 and 96.5 % similarity, respectively). The level of DNA–DNA relatedness between strains WSY08-1T and A. halophilus B6BT was 31.4 %. On the basis of its phenotypic, chemotaxonomic and genotypic characteristics, strain WSY08-1T is considered to represent a novel species in the genus Aquibacillus, for which the name Aquibacillus salifodinae sp. nov. is proposed. The type strain is WSY08-1T (=JCM 19761T = CGMCC 1.12849T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Aquibacillus was proposed by Amoozegar et al. (2014) with the description of Aquibacillus halophilus as well as the transfer of Virgibacillus koreensis to Aquibacillus koreensis and Virgibacillus albus to Aquibacillus albus. Therefore, at the time writing, this genus, belonging to the family Bacillaceae, comprises three species with validly published names and the type species is A. halophilus. The type strains of all three species were isolated from saline environments. Members of the genus Aquibacillus are Gram-stain positive, rod-shaped, motile, produce endospores, moderately halophilic, catalase- and oxidase-positive. Chemotaxonomically they are characterized by having MK-7 as the predominant respiratory quinone, phosphatidylglycerol and diphosphatidylglycerol as the major polar lipids, anteiso-C15:0 as the major fatty acid and meso-diaminopimelic acid (meso-DAP) as the diagnostic diamino acid in the peptidoglycan. The DNA G+C content of members of this genus ranges from 35.8–41.0 mol%.
During our surveys on bacterial diversity of a salt mine in Wensu county, Akesu area in Xinjiang province of China, a bacterial strain named WSY08-1T was obtained. By using a polyphasic taxonomic approach, we conclude that strain WSY08-1T represents a novel species of the genus Aquibacillus, for which the name Aquibacillus salifodinae sp. nov. is proposed.
Materials and methods
Bacterial strains and culture condition
Strain WSY08-1T was isolated from a salt mine sample (salt crystal, 41°34′52″N, 80°43′37″E) in Xinjiang province, China. Marine broth 2216 (MB, Difco) was used for isolation. The medium was solidified with 2.0 % agar (MA). To enrich bacteria about 3 g salt sample was suspended in 30 ml MB at 28 °C for 2 days. The enrichment culture was diluted and spread onto MA plates which were incubated at 28 °C. After 3 days of incubation, one of the aerobic isolates, strain WSY08-1T, was observed to produce cream-pigmented, circular and elevated colonies on MA plates. One of these colonies was picked up and purified by repeated restreaking. The isolate was routinely cultured on MA plates and maintained at −80 °C with 25 % (v/v) glycerol.
Biomass for chemotaxonomic and molecular studies was obtained by cultivation in shaking flasks with MB at 28 °C for 2 days. The recommended Minimal Standards for describing new taxa of aerobic, endospore-forming bacteria were followed (Logan et al. 2009). The reference type strain A. halophilus B6BT were obtained from Korean Collection for Type Cultures (KCTC; Korea); A. koreensis JCM 12387T and A. albus JCM 17364T were obtained from the Japan Collection of Microorganisms (JCM; Japan). Reference type strains were cultured under the same conditions as strain WSY08-1T for comparative analyses.
Morphological, physiological and biochemical characterization
Cell morphology was determined by optical microscopy (BX40; Olympus) and transmission electron microscopy (JEM-1230; JEOL) (Huo et al. 2010). Motility was detected by optical microscopy using the hanging drop technique (Skerman 1967). Gram staining was performed by following the method outlined by Dussault (1955). To determine the growth conditions of strain WSY08-1T, MB-1 medium with various NaCl concentrations (0–15 %, at intervals of 1 %, w/v) was used. MB-1 medium contained (per l distilled water): peptone (Becton–Dickinson) 5.0 g, yeast extract (Becton–Dickinson) 1.0 g, Ferric citrate 0.1 g, MgCl2·6H2O 12.6 g, MgSO4·7H2O 6.64 g, CaCl2 1.8 g, KCl 0.55 g, KBr 0.08 g, SrCl 34.0 mg, Boric acid 22.0 mg and NH4NO3 2.4 mg. The ranges of pH and temperature for growth were determined by using MB medium according to Zhang et al. (2013).
Catalase and oxidase activities and nitrate and nitrite reduction were tested according to (Dong and Cai 2001). Indole, methyl red and Voges-Proskauer tests, H2S production, as well as hydrolysis of aesculin, casein, gelatin and starch were performed as described by Zhu (2011). Esterase activity was detected as outlined by (Gutiérrez and González 1972). To analyze the use of accessory electron acceptors, sodium thiosulfate (20 mM), sodium sulfate (20 mM), sodium sulfite (5 mM), sodium nitrite (5 mM) and sodium nitrate (20 mM) were respectively added to sterile MB. Oxygen was removed as described by Grishchenkov et al. (2000). The same medium lacking l-cysteine and resazurin was used as aerobic control. Other enzyme activities and physiological properties were determined by using API ZYM, API 20E and API 20NE kits, using distilled water supplemented with 4 % (w/v) NaCl to suspend the cells. Acid production tests were carried out by using API 50CH kits, using modified MB from which yeast extract and peptone were omitted and 0.05 g/L yeast extract was added. Utilization of single carbon sources for growth was performed using Biolog GP2 96-well Microplates (Biolog) by using the modified MB. All API tests were performed according to the manufacturer’s instructions (bioMérieux). Antibiotic sensitivity tests were determined on MA plates with antibiotic discs containing the following amount (μg per disc, unless indicated): amikacin (30), ampicillin (10), bacitracin (0.04 IU), chloramphenicol (30), ciprofloxacin (5), erythromycin (15), gentamicin (10), kanamycin (30), neomycin (30), norfloxacin (10), novobiocin (30), nystatin (100), penicillin G (10 IU), polymyxin (300 IU), rifampicin (5), streptomycin (10), sulfamethoxazole (300), tobramycin (10) and vancomycin (30).
Chemotaxonomic characterization
For chemotaxonomic experiments, late exponential phase cells of strains WSY08-1T, A. koreensis JCM 12387T and A. albus JCM 17364T were harvested from MB. The isomer type of the diamino acid in the cell-wall peptidoglycan and isoprenoid quinones were determined as described by Komagata and Suzuki (1987). Polar lipids of strain WSY08-1T and reference strains were extracted and separated on silica gel 60 F254 aluminium-backed thin-layer plates (10 × 10 cm, Merk 5554) and further analyzed as described by Minnikin et al. (1984). Fatty acid methyl esters were obtained from freeze-dried cells as described by Kuykendall et al. (1988) and their identification and quantification were performed using the Sherlock Microbial Identification System (MIDI) with the standard MIS Library Generation Software version 4.5 (Microbial ID).
Molecular studies
The 16S rRNA gene was amplified and analyzed as described previously (Xu et al. 2007). PCR products were cloned into vector pMD19-T (TaKaRa) and then sequenced. The sequence was compared with closely related sequences of reference species using the EzTaxon-e server (Kim et al. 2012) and BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple sequences were aligned with CLUSTAL W1.8 (Thompson et al. 1994). Phylogenetic trees were constructed by the neighbour-joining (Saitou and Nei 1987), the maximum-parsimony (Fitch 1971) and the maximum-likelihood (Felsenstein 1981) methods within the MEGA 5 program package (Tamura et al. 2011). Evolutionary distances were calculated according to the algorithm of Kimura’s two-parameter model (Kimura 1980) for the neighbour-joining method.
The DNA G+C content was determined by reversed-phase HPLC as described by Mesbah and Whitman (1989). DNA–DNA hybridizations were performed by using the thermal denaturation and renaturation method (De Ley et al. 1970) as modified by Huß et al. (1983), using a Beckman DU 800 Spectrophotometer. The experiments were carried out in quadruplicate.
Results and conclusion
Morphological, physiological and biochemical characteristics
Colonies of strain WSY08-1T on MA plates after 2 days incubation were observed to be 0.1–0.3 mm in diameter, cream-pigmented, circular and elevated. Strain WSY08-1T was found to be strictly aerobic. Cells were observed to be Gram-stain positive, rod-shaped (0.4–0.6 × 5.0–8.1 μm), motile by means of peritrichous flagella and endospore-forming. Spherical to ellipsoidal endospores were observed to be formed terminal in swollen sporangia (Fig. S1). The physiological and biochemical characteristics of strain WSY08-1T that differentiate it from the reference strains are shown in Table 1. The detailed physiological and biochemical characteristics of the type strain WSY08-1T are given in the species description.
The type strain was found to be sensitive to ampicillin, bacitracin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, kanamycin neomycin, norfloxacin, novobiocin, penicillin G, rifampicin, sulfamethoxazole, tobramycin and vancomycin; and resistant to amikacin, nystatin, polymyxin and streptomycin.
Chemotaxonomy results
The cell wall peptidoglycan of strain WSY08-1T was found to contain meso-diaminopimelic acid as the diamino-acid. The isoprenoid quinones were identified as MK-7 (94 %) and MK-6 (6 %). The polar lipids were found to include phosphatidylglycerol, diphosphatidylglycerol, an unknown glycolipid, two unknown phospholipids and an unknown lipid (Fig. S2). The major fatty acids were identified as anteiso-C15:0 (72.7 %) and anteiso-C17:0 (10.0 %). The complete fatty acid profiles of strain WSY08-1T and the reference type strains are summarized in Table 2.
Molecular characterization
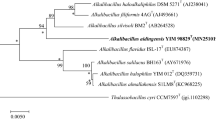
The nearly complete 16S rRNA gene sequence (1,528 bp) of strain WSY08-1T was determined (GenBank accession number AB859945). Based on 16S rRNA gene sequence analysis, the validly named species that shared highest sequence similarities to strain WSY08-1T were identified as A. halophilus B6BT (97.6 %), followed by A. koreensis BH30097T (96.9 %), A. albus YIM 93624T (96.5 %), Sediminibacillus halophilus En8dT (96.4 %) and Virgibacillus carmonensis JCM 16508T (96.0 %). Type strains of other species shared lower sequence similarities (<96 %) to strain WSY08-1T. Phylogenetic trees revealed that strain WSY08-1T clustered with A. halophilus B6BT, A. koreensis BH30097T and A. albus YIM 93624T in a separate clade (Fig. 1; Fig. S3 and Fig. S4). DNA–DNA hybridization between strains WSY08-1T and A. halophilus B6BT showed a low relatedness value of 31.4 %.
Maximum-likelihood phylogenetic tree based on 16S rRNA gene sequences showed the relationships of strain WSY08-1T and related species. Bootstrap values based on 1,000 replications are listed as percentages at branching points. Bootstrap values higher than 50 % are indicated at branch-points. Enterococcus faecium JCM 5803T was used as outgroup. Bar 1 % sequence divergence
The genomic G+C content of strain WSY08-1T was determined to be 36.9 mol%, which is in the range for the members of the genus Aquibacillus.
Taxonomic conclusion
Based on its characterisation as Gram-positive, rod-shaped, endospore-forming, moderately halophilic cells; positive for catalase and oxidase activities; the major fatty acid (anteiso-C15:0); the major polar lipids (phosphatidylglycerol and diphosphatidylglycerol); the predominant menaquinone (MK-7); the cell-wall peptidoglycan diamino acid (meso-diaminopimelic acid); the G+C content (36.9 mol%); and the phylogenetic analyses, all these data suggest that strain WSY08-1T should belong to the genus Aquibacillus. However, strain WSY08-1T showed notable differences in comparison to the type strains of the most closely related species A. halophilus B6BT, A. koreensis BH30097T and A. albus YIM 93642T with regard to morphological characteristics, growth ranges (temperature, pH and NaCl tolerance), nitrate reduction, Voges–Proskauer, hydrolysis of starch and Tween 80, and acid production from carbohydrates (Table 1). The cellular fatty acid profile of strain WSY08-1T was qualitatitively similar to that of the reference strains but differences in the proportions of some fatty acids were noted (Table 2). Differences in the polar lipid profile also existed compared to the reference strains (Fig. S4). Strain WSY08-1T shared low 16S rRNA gene sequence similarities to the type strains A. halophilus B6BT, A. koreensis BH30097T and A. albus YIM 93624T (97.6, 96.9 and 96.5 %, respectively). DNA–DNA hybridization data (31.4 %) clearly differentiated strain WSY08-1T from strain A. halophilus B6BT.
Based on the phenotypic, chemotaxonomic and genotypic characteristics described above, strain WSY08-1T was identified as a novel species within the genus Aquibacillus, for which the name Aquibacillus salifodinae sp. nov. is proposed.
Description of Aquibacillus salifodinae sp. nov
Aquibacillus salifodinae (sa.li.fo.di’nae. L. n. sal, salt; L. n. fodina, a pit; N.L. gen. n. salifodinae of a saltpit, salt mine).
Cells are Gram-positive, strictly aerobic, motile by means of peritrichous flagella and rod-shaped (0.4–0.6 × 5.0–8.1 μm). Spherical to ellipsoidal endospores are formed in terminal swollen sporangia. Colonies are 0.1–0.3 mm in diameter, cream-pigmented, circular and elevated. Growth occurs at 20–45 °C (optimum 37 °C), 0–10 % (w/v) NaCl (optimum 4 %, w/v) and pH 6.0–9.0 (optimum 7.0). Catalase and oxidase activities are positive. Nitrate is reduced to nitrite but not to nitrogen. Indole, methyl red, Voges–Proskauer, H2S production, hydrolysis of starch, casein and gelatin, as well as the production of arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase and urease are negative. Clearly positive in the Biolog GP2 MicroPlate system for the oxidation of acetic acid, N-acetylglucosamine, N-acetyl-l-glurtamic acid, adenosine-5′-monophosphate, l-alaninamide, l-alanyl-glycine, amygdalin, l-asparagine, l-arabinose, arbutin, d-cellobiose, α-cyclodextrin, 2′-deoxy adenosine, l-fructose, d-fructose, d-fructose-6-phosphate, d-galactose, gentiobiose, d-gluconic acid, d-glucose, α-d-glucose-1-phosphate, d-glucose-6-phosphate, l-glutamic acid, d-l-α-glycerol phosphate, glycyl-l-glutamic acid, glycerol, glycogen, γ-hydroxybutyric acid, m-inositol, inosine, inulin, α-ketoglutaric acid, α-ketovaleric acid, lactamide, α-d-lactose, lactulose, d-malic acid, l-malic acid, maltose, mannan, d-mannose, d-melezitose, d-melibiose, α-methyl-d-galactoside, β-methyl-d-galactoside, 3-methyl-glucose, α-methyl-d-glucoside, α-methyl-d-mannoside, palatinose, propionic acid, d-psicose, pyruvic acid, l-pyroglutamic acid, d-raffinose, l-rhamnose, d-ribose, salicin, sedoheptulosan, d-sorbitol, succinamic acid, succinic acid, succinic acid mono-methyl ester, sucrose, d-tagatose, thymidine, thymidine-5′-monophosphate, trehalose, Tween 40 and 80, uridine-5′-monophosphate, xylitol and d-xylose. Other substrates in the GP2 MicroPlate are not utilized as sole carbon source. According to the API 50 CH system, positive for acid production from N-acetylglucosamine, d-adonitol, aesculin, amygdalin, l-arabinose, l-arabitol, arbutin, cellobiose, d-fructose, gentiobiose, d-glucose, 2-ketogluconate, 5-ketogluconate, d-lyxose, maltose, d-mannose, β-methyl-d-xylopyranoside, d-ribose, salicin, sucrose, trehalose and d-xylose; Weakly positive of glycerol, l-sorbose, d-tagatose and xylitol; other substrates of the API 50 CH system are not converted to acid. With API ZYM systems, alkaline phosphatase, esterase (C4), esterase lipase (C8), β-glucosidase, β-glucuronidase, naphtol-AS-BI-phosphohydrolase are positive; acid phosphohydrolase is weakly positive; lipase (C14), leucine arylamidase, valine arylamidase, cysteine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, α-glucosidase. N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase are negative. The cell-wall peptidoglycan contains meso-diaminopimelic acid as the diamino-acid. MK-7 is the predominant menaquinone and MK-6 is also present. The polar lipids are phosphatidylglycerol, diphosphatidylglycerol, an unknown glycolipid, two unknown phospholipids and an unknown lipid. The major fatty acids are anteiso-C15:0 and anteiso-C17:0. The DNA G+C content of the type strain is 36.9 mol% (as determined by HPLC).
The type strain WSY08-1T (= JCM 19761T = CGMCC 1.12849T) was isolated from a salt mine taken from Wensu county, Xinjiang province, China. The GenBank accession number of the 16S rRNA gene sequence of the type strain is AB859945.
References
Amoozegar MA, Bagheri M, Didari M, Mehrshad M, Schumann P, Spröer C, Sánchez-Porro C, Ventosa A (2014) Aquibacillus halophilus gen. nov., sp. nov., a moderately halophilic bacterium from a hypersaline lake, and reclassification of Virgibacillus koreensis as Aquibacillus koreensis comb. nov. and Virgibacillus albus as Aquibacillus albus comb. nov. Int J Syst Evol Microbiol 64:3616–3623
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Dong XZ, Cai MY (2001) Determination of biochemical properties. In: Dong XZ, Cai MY (eds) Manual for the systematic identification of General Bacteria. Science Press, Beijing, pp 370–398 in Chinese
Dussault HP (1955) An improved technique for staining red halophilic bacteria. J Bacteriol 70:484–485
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Grishchenkov VG, Townsend RT, McDonald TJ, Autenrieth RL, Bonner JS, Boronin AM (2000) Degradation of petroleum hydrocarbons by facultative anaerobic bacteria under aerobic and anaerobic conditions. Process Biochem 35:889–896
Gutiérrez C, González C (1972) Method for simultaneous detection of proteinase and esterase activities in extremely halophilic bacteria. Appl Microbiol 24:516–517
Huo YY, Xu XW, Cui HL, Wu M (2010) Gracilibacillus ureilyticus sp. nov., a halotolerant bacterium from a saline-alkaline soil. Int J Syst Evol Microbiol 60:1383–1386
Huß VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Method Microbiol 19:161–207
Kuykendall LD, Roy MA, O’Neill JJ, Devine TE (1988) Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Bacteriol 38:358–361
Lee JS, Lim JM, Lee KC, Lee JC, Park YH, Kim CJ (2006) Virgibacillus koreensis sp. nov., a novel bacterium from a salt field, and transfer of Virgibacillus picturae to the genus Oceanobacillus as Oceanobacillus picturae comb. nov. with emended descriptions. Int J Syst Evol Microbiol 56:251–257
Logan JM, Berge O, Bishop AH, Busse HJ, De Vos P, Fritze D, Heyndrickx M, Kämpfer P, Rabinovitch L, Salkinoja-Salonen MS, Seldin L, Ventosa A (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59:2114–2121
Mesbah M, Whitman WB (1989) Measurement of deoxyguanosine/thymidine ratios in complex mixtures by high-performance liquid chromatography for determination of the mole percentage guanine + cytosine of DNA. J Chromatogr 479:297–306
Minnikin DE, Odonnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Method 2:233–241
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Skerman VBD (1967) A guide to the identification of the genera of bacteria, 2nd edn. Williams & Wilkins, Baltimore
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. doi:10.1093/molbev/msr121
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Xu XW, Wu YH, Zhou Z, Wang CS, Zhou YG, Zhang HB, Wang Y, Wu M (2007) Halomonas saccharevitans sp. nov., Halomonas arcis sp. nov., and Halomonas subterranea sp. nov., halophilic bacteria isolated from hypersaline environments of China. Int J Syst Evol Microbiol 57:1619–1624
Zhang YJ, Zhou Y, Ja M, Shi R, Chun-Yu WX, Yang LL, Tang SK, Li WJ (2012) Virgibacillus albus sp. nov., a novel moderately halophilic bacterium isolated from Lop Nur salt lake in Xinjiang province, China. Antonie van Leeuwenhoek 102:553–560
Zhang WY, Fang MX, Zhang WW, Xiao C, Zhang XQ, Yu ZP, Zhu XF, Wu M (2013) Extensimonas vulgaris gen. nov., sp. nov., a novel member of the family Comamonadaceae. Int J Syst Evol Microbiol 63:2062–2068
Zhu XF (2011) Modern experimental technique of microbiology. Zhejiang University Press, Hangzhou English translation
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 31170001).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, WY., Hu, J., Zhang, XQ. et al. Aquibacillus salifodinae sp. nov., a novel bacterium isolated from a salt mine in Xinjiang province, China. Antonie van Leeuwenhoek 107, 367–374 (2015). https://doi.org/10.1007/s10482-014-0335-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0335-9