Abstract
Two Gram-negative, moderately thermophilic, yellow-pigmented, rod-shaped and motile bacterial strains, designated YIM 75003T and YIM 75004T, were isolated from sediment samples collected from the Tagejia hot spring in Tibet, western China. The taxonomic affiliation of the two strains was investigated by a polyphasic approach. Pairwise comparison of the 16S rRNA gene sequences showed that strains YIM 75003T and YIM 75004T are closely related to Altererythrobacter buctense M0322T (97.2 and 97.1% respectively), while sharing 99.9% sequence similarity against each other. Optimum growth of the two strains was observed at 37–45 °C, pH 8.0 and in the presence of 1–6% NaCl (w/v). Their predominant respiratory quinone was found to be ubiquinone 10. The major fatty acids in both the strains were identified as summed feature 8 (C18:1 ω6c and/or C18:1 ω7c) and summed feature 4 (C17:1 anteiso B and/or iso I). Their major polar lipid profiles were found to be diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol and sphingoglycolipid. The DNA G+C contents of strains YIM 75003T and YIM 75004T were determined to be 61.3 and 60.1 mol%, respectively. The DNA-DNA hybridization values between strains YIM 75003T and YIM 75004T, and between the two strains and their closely related phylogenetic neighbour A. buctense M0322T (=CGMCC 1.12871T) were less than 70%. Based on the morphological and physiological properties, phylogenetic analyses, chemotaxonomic characteristics and DNA–DNA relatedness values, the two strains can be distinguished from each other and from their phylogenetically closely related strain. Strains YIM 75003T and YIM 75004T are, therefore, considered to represent two novel species of the genus Altererythrobacter, for which the names Altererythrobacter lauratis sp. nov. (type strain YIM 75003T = KCTC 52606T = CCTCC AB2016268T) and Altererythrobacter palmitatis sp. nov. (type strain YIM 75004T = KCTC 52607T = CCTCC AB2016270T) are proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hot springs are one of the extreme ecosystems with limited life forms due to their harsh conditions. Despite the extreme environments, Tibet’s hot springs have been reported to possess diverse microbial communities, including several novel taxa (Huang et al. 2011). Microorganisms from such extreme habitats may be an important source for novel enzymes and metabolites applicable to biotechnological industries (Song et al. 2009). The genus Altererythrobacter, belonging to the family Erythrobacteraceae, was first described by Kwon et al. (2007), and emended subsequently by Xue et al. (2012, 2016). At the time of writing, the genus contains 25 species with validly published names, the majority of which were isolated from deep-sea sediment (e.g. Wu et al. 2014; Zhang et al. 2016a) or sea water (e.g. Park et al. 2011). Phylogenetically, the genus is polyphyletic in nature with members of this genus being grouped along with the genera Erythrobacter and Porphyrobacter within the family Erythrobacteraceae. The characteristic features of this genus include Gram-staining negative, absence of bacteriochlorophyll a (Bchl a) and Q-10 as dominant respiratory quinone. Only four species have been reported to possess flagella (Seo and Lee 2012; Nedashkovskaya et al. 2013; Wu et al. 2014; Zhao et al. 2017). During an investigation of microbial diversity of hot springs, two yellow-pigmented, moderately thermophilic strains were isolated from samples collected from the Tagejia geothermal fields of the Tibetan Plateau. This paper reports the taxonomic characterization of the two strains YIM 75003T and YIM 75004T as novel species of the genus Altererythrobacter.
Materials and methods
Strains and culture conditions
Strains YIM 75003T and YIM 75004T were isolated from samples collected from the Tagejia geothermal field (29°24′51.11″N, 86°43′58.02″E) by serial dilution plating on Cellulose-Casamino (CC) agar (Microcrystalline cellulose, 1.0 g; Casamino acid, 1.0 g; KNO3, 0.2 g; Na2HPO3, 0.5 g; MgSO4, 0.05 g; FeSO4, 0.01 g; Agar, 12.0 g; Distilled water, 1 L; pH 7.2 ± 0.2). The isolation plates were kept incubated at 45 °C for 15 days. Single colonies were repeatedly streaked on Reasoner’s 2A agar (R2A agar; Reasoner and Geldreich 1985) for purification, and pure cultures were routinely cultivated on the same medium. Both strains were stored as glycerol suspensions (20%, v/v) at −80 °C. The type strain Altererythrobacter buctense M0322T (=CGMCC 1.12871T) was obtained from the China General Microbiological Culture Collection Center (CGMCC) and was cultivated on R2A agar at 30 °C for comparative taxonomic work.
Morphological, physiological and biochemical characterization
For morphological studies, strains YIM 75003T and YIM 75004T were cultured at 45 °C on R2A agar adjusted to pH 7.2. Morphology was examined by light microscopy (model BH2; Olympus) and transmission electron microscopy (JEM-2100; JEOL). Gram staining was carried out by using the standard Gram staining procedure and confirmed by KOH lysis test (Buck 1982). Growth at different temperatures (4, 10, 15, 20, 28, 37, 45, 50, 55, 60, 65 and 70 °C) and NaCl tolerance (0–12.0% w/v, at intervals of 1.0 unit) was determined using R2A broth. The pH range (4.0–11.0, at intervals of 1.0 pH unit) for growth was tested in R2A broth using the buffer system described by Xu et al. (2005). Growth on several media such as nutrient agar (NA), trypticase soy agar (TSA) (Waksman 1967), Luria–Bertani (LB) agar (Atlas 1997), R2A agar, MR agar (Zhang et al. 2016b) and other media at 45 °C were also evaluated. Oxidase activity was tested via the oxidation of tetramethyl-p-phenylenediamine (Kovacs 1956). Catalase activity was detected by assessing the production of bubbles on addition of a drop of 3% (v/v) H2O2. H2S production, milk peptonization and coagulation, urease activity and hydrolysis of cellulose, gelatin, starch and Tweens (20, 40 and 80) were performed as described by Gonzalez et al. (1978). Sole carbon source utilization was determined using the methods described by Shirling and Gottlieb (1966) and Locci (1989), while sole nitrogen source utilization was examined according to Williams et al. (1989). Cellular pigments were extracted as described by Rainey et al. (2003) using culture grown on R2A broth under darkness, and absorbance of the pigments were measured with a U-4100 spectrophotometer (HITACHI, Japan). Other biochemical tests were carried out using the API 20NE, API ZYM and API 50CHB/E test strips (bioMérieux) according to the manufacturer’s instructions. Antibiotic susceptibility tests were performed by the agar-diffusion method on R2A agar (30 °C, 4 days) after plating with bacterial suspensions equivalent to 0.5 McFarland standards.
Chemotaxonomy
The polar lipids were prepared as described by Minnikin et al. (1979) and identified by two-dimensional TLC (Collins and Jones 1980). Menaquinones were extracted (Collins et al. 1977) and analysed using HPLC (Kroppenstedt 1982). Cellular fatty acids analysis was performed by using the Microbial Identification System (Sherlock Version 6.1; MIDI database: TSBA6; Sasser 2001). Biomass for fatty acid analysis was obtained from cells grown on R2A agar at 30 °C for 4 days. The G+C contents of the genomic DNAs were determined by using reversed-phase HPLC (Mesbah et al. 1989), with Escherichia coli DH5α as the reference strain.
Molecular analysis and DNA–DNA hybridization
Extraction of genomic DNAs and PCR amplification of the 16S rRNA genes were performed as described by Li et al. (2007). The amplicons were purified, cloned into Trans1-T1 chemically competent cells using pEASY-T1 vector and sequenced by Sangon Biotech (Shanghai). The sequences were compared with available 16S rRNA gene sequences of validly published species from the EzBioCloud server (Yoon et al. 2016). Multiple alignments with sequences of the most closely related taxa were carried out by using CLUSTAL_X program (Thompson et al. 1997). Phylogenetic analyses were performed by using three tree-making algorithms: neighbour–joining (Saitou and Nei 1987), maximum-likelihood (Felsenstein 1981) and maximum-parsimony (Fitch 1971). The dendrograms were generated by using the MEGA version 6.0 software package (Tamura et al. 2013). Kimura’s two parameter model (Kimura 1980) was used to calculate evolutionary distance matrices of the neighbour–joining tree. Bootstrap analysis was performed with 1000 replications (Felsenstein 1985). Caulobacter vibrioides CB51T (AJ009957) was used as outgroup. DNA–DNA relatedness was studied between strains YIM 75003T and YIM 75004T, and with the phylogenetically closely related strain A. buctense M0322T (=CGMCC 1.12871T) using the fluorometric micro-well method (Ezaki et al. 1989; Christensen et al. 2000) at 45 °C as the optimal hybridization temperature. One of the two DNA molecules for hybridization was labeled while the other was immobilized, and vice versa. Six replications were done for each sample and the two extreme values (highest and lowest) for each sample were excluded. The relatedness values are expressed by calculating the means of the remaining four values.
Results and discussion
Phenotypic characteristics
Cells of strains YIM 75003T and YIM 75004T were observed to be Gram-negative, aerobic, rod-shaped and motile. Transmission electron microscopy showed that strain YIM 75003T measured 0.3–0.7 μm in width and 1.2–3.8 μm in length, while strain YIM 75004T measured about 0.3–0.7 μm in width and 0.8–1.6 μm in length (Fig. S1). Both the strains possessed a polar flagellum (Fig. S1). Colonies of the two strains on R2A agar after 3 days of incubation at 45 °C were yellow-pigmented. While colonies of strain YIM 75003T were smooth and measured approximately 1.5–2 mm in diameter, those of strain YIM 75004T were found to be rough and approximately 0.6–0.8 mm in diameter. Both strains were found to grow well on R2A and MR agar, but not on LB agar, TSA or NA. Growth was observed at a temperature range of 10–55 °C (optimum, 37–45 °C) and in the presence of up to 11% NaCl (w/v) (optimum, 1–6%). The two strains showed optimum growth at pH 8.0 but differed in their pH range for growth. Strain YIM 75003T exhibited growth at pH 5.0–11.0 while strain YIM 75004T at pH 5.0–9.0. The two strains were observed to be positive for catalase, oxidase and cellulase activities, but negative for H2S production and urease activity. The two strains utilised acetate sodium, citrate sodium, fucose, glycerol, inositol, malate sodium, maltose, mannitol, mannose, succinate sodium, sucrose, alanine, arginine, cysteine, glycine, isoleucine, ornithine, phenylalanine, proline, serine, tyrosine, threonine and valine as sole carbon or nitrogen sources, but not arabinose, fructose, galactose, glucose, glycine, rhamnose or xylose. While strain YIM 75003T utilised pyruvate sodium and saccharose, strain YIM 75004T could utilise ribose, sorbose and xylitol. Other biochemical activities as determined by the API 20NE, API ZYM and API 50CHB/E test strips (bioMérieux) are presented in Supplementary Table S1. The pigment extracts of the two strains showed absorption maxima at 453 and 480 nm, and did not exhibit any characteristic absorbance at 805 and 830–890 nm indicating the absence of Bchl a. The two strains were found to be sensitive to amikacin (30 μg), cefuroxime sodium (30 μg), chloramphenicol (30 μg), erythromycin (15 μg), gentamicin (10 μg), penicillin (10 IU), piperacillin (100 μg) polymyxin B (300 IU), sulfamethoxazole (300 μg), tetracycline (30 μg) and vancomycin (30 μg), but resistant to ethylhydrocupreine (5 μg). Furthermore, strain YIM 75004T was sensitive to ciprofloxacin (5 μg) and oxacillin (1 μg), but not strain YIM 75003T. Detailed physiological characteristics of the two strains are given in the species description, and the differential characteristics between strains YIM 75003T, YIM 75004T and the closely related type strains of the genus Altererythrobacter are listed in Table 1.
Chemotaxonomy
The major respiratory quinone identified in strains YIM 75003T and YIM 75004T was observed to be ubiquinone-10 (Q-10), however small percentages of Q-8 and Q-9 were also detected. The major fatty acids (>10%) of strains YIM 75003T were summed feature 8 (C18:1 ω6c and/or C18:1 ω7c), summed feature 4 (C17:1 anteiso B and/or iso I) and C16:0, while for strain YIM 75004T were summed feature 8 (C18:1 ω6c and/or C18:1 ω7c), summed feature 4 (C17:1 anteiso B and/or iso I), C17:1 ω6c and C16:0 (Table 2). The major polar lipid profiles of strains YIM 75003T and YIM 75004T consisted of diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol and sphingoglycolipid (Fig. S2). While two unidentified glycolipids and four unidentified lipids were also detected in the former as minor polar lipids, two unidentified glycolipids and two unidentified lipids were found in the latter (Fig. S2). The G+C contents of strains YIM 75003T and YIM 75004T were determined to be 61.3 and 60.1 mol%, respectively.
Phylogenetic analysis and DNA–DNA relatedness value
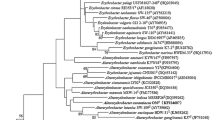
Sequence comparison of the almost complete 16S rRNA gene sequences of strains YIM 75003T (GenBank Accession Number KX808673) and YIM 75004T (KX808674) using the NCBI-BlastN analyses showed that the two strains are closely related to members of the genus Altererythrobacter and showed high sequence similarities with A. buctense M0322T (97.2 and 97.1%, respectively). The two strains however showed 99.9% sequence similarity among themselves. Phylogenetic trees based on 16S rRNA gene sequences were generated using the 1343 unambiguously aligned positions that remained after trimming in the MEGA 6 software. In the neighbour–joining phylogenetic tree (Fig. 1), the two strains formed a close clade with A. buctense M0322T, A. salegens XY-R17T and A. atlanticus 26DY36T. The stability of the neighbour–joining tree was supported by the dendrograms generated with maximum likelihood and maximum parsimony methods (Figs. S3 and S4). Since the sequence similarities among the two strains and with A. buctense M0322T were above 97%, DNA–DNA hybridization experiments were performed between strains YIM 75003T, YIM 75004T and A. buctense M0322T (= CGMCC 1.12871T) to determine their genomic DNA relatedness. The DNA–DNA relatedness values of strains YIM 75003T and YIM 75004T with A. buctense M0322T (=CGMCC 1.12871T) was determined to be 53.0 ± 2.2 and 41.5 ± 1.1%, respectively, while the two strains had a relatedness value of 44.8 ± 1.4% between themselves (Table S2). These values are less than cut-off point (70%) for the delineation of genomic species (Wayne et al. 1987).
Neighbor–joining phylogenetic tree based on 16S rRNA gene sequences, showing the position of strains YIM 75003T and YIM 75004T among related species of the family Erythrobacteraceae. The sequence of Caulobacter vibrioides CB51T/AJ009957 was used as outgroup. Asterisks indicate branches of the tree that were also found in dendrograms generated with the maximum-likelihood and maximum-parsimony algorithms. Numbers on branch nodes are bootstrap values (1000 resamplings, only values over 50% are given). Bar represents 0.02 substitutions per nucleotide position
Based on the phylogenetic analysis (Fig. 1) and comparison of the phenotypic and chemotaxonomic markers such as presence of Q-10 as respiratory quinone (Fig. S2), strains YIM 75003T and YIM 75004T should be affiliated to the genus Altererythrobacter. The major polar lipid profiles of the two strains were found to be similar to that of A. salegens XY-R17T (Liang et al. 2016), but can be differentiated from the closely related type strain A. buctense M0322T by the presence of diphosphatidylglycerol and sphingoglycolipid (Zhang et al. 2016b). Other characteristics that differentiate the two strains from related type strains of the genus Altererythrobacter included differences in growth conditions (temperature and pH ranges for growth), utilization of carbon and nitrogen sources, as well as the proportions of some fatty acids (Tables 1, 2). Notably the two strains can also be differentiated from each other in the hydrolysis of Tweens 20 and 40, nitrate reduction, utilization of pyruvate, ribose, saccharose, sorbose, xylitol, histidine and lysine, activities of cystine arylamidase and leucine arylamidase, and acid production from d-fructose. In addition, the DNA–DNA relatedness value between them is moderately low, indicating that the two strains YIM 75003T and YIM 75004T merit classification as two novel species within the genus Altererythrobacter, for which the names Altererythrobacter lauratis sp. nov. and Altererythrobacter palmitatis sp. nov. are proposed, respectively.
Description of Altererythrobacter lauratis sp. nov.
Altererythrobacter lauratis [lau.ra’tis. N.L. n. lauras, atis, laurate (chemical); N.L. gen. n. lauratis of laurate, because it oxidises laurate].
Cells are Gram-negative, aerobic, motile and short rods (approximately 0.3–0.7 × 1.2–3.8 μm). Colonies on R2A agar are smooth, convex, circular and yellow-pigmented. Growth occurs on R2A and MR agar but not on LB agar, NA or TSA. Growth occurs at 10–55 °C (optimum, 37–45 °C), pH 5.0–11.0 (optimum, pH 8.0) and 0–11% NaCl (w/v) (optimum, 1–6%). Does not contain Bchl a as a photosynthetic pigment. Positive for catalase and oxidase activities. Hydrolyses cellulose and Tween 20, but not chitin, starch or Tweens 40 and 80. H2S is not produced. The respiratory quinone is Q-10. The polar lipids are diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, sphingoglycolipid, two unidentified glycolipids and four unidentified lipids. The major fatty acids (>10%) are summed feature 8 (C18:1 ω6c and/or C18:1 ω7c), summed feature 4 (C17:1 anteiso B and/or iso I) and C16:0.
The type strain, YIM 75003T (=KCTC 52606T = CCTCC AB2016268T), was isolated from the hot spring of Tagejia geothermal field in Tibet, western China. The DNA G+C content of the type strain is 61.3%. The GenBank accession number for the 16S rRNA gene sequence of strain YIM 75003T is KX808673. The taxon number of the strain in the Digital Protologue database is TA00084.
Description of Altererythrobacter palmitatis sp. nov.
Altererythrobacter palmitatis [pal.mi.ta’tis. N.L. n. palmitas, atis palmitate (chemical); N.L. gen. n. palmitatis of palmitate, because it oxidises palmitate].
Cells are Gram-negative, aerobic, motile and short rods (approximately 0.3–0.7 × 0.8–1.6 μm). Colonies on R2A agar are dry, convex, circular and yellow-pigmented. Grows well on R2A and MR agar but not on LB agar, NA or TSA. Growth occurs at 10–55 °C (optimum, 37–45 °C), pH 5.0–9.0 (optimum, pH 8.0) and 0–11% NaCl (w/v) (optimum, 1–6%). Does not contain Bchl a as a photosynthetic pigment. Positive for catalase and oxidase activities. Hydrolyses cellulose and Tween 40, but not chitin, starch or Tweens 20 and 80. H2S is not produced. The respiratory quinone is Q-10. The polar lipids are diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, sphingoglycolipid, two unidentified glycolipids and two unidentified lipids. The major fatty acids (>10%) are summed feature 8 (C18:1 ω6c and/or C18:1 ω7c), summed feature 4 (C17:1 anteiso B and/or iso I), C17:1 ω6c and C16:0.
The type strain, YIM 75004T (=KCTC 52607T = CCTCC AB2016270T), was isolated from the hot spring of Tagejia geothermal field in Tibet, western China. The DNA G+C content of the type strain is 60.1%. The GenBank Accession Number for the 16S rRNA gene sequence of strain YIM 75004T is KX808674. The taxon number of the strain in the Digital Protologue database is TA00085.
References
Atlas RM (1997) Handbook of microbiological media, 2nd edn. CRC Press, New York
Buck JD (1982) Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993
Christensen H, Angen O, Mutters R, Olsen JE, Bisgaard M (2000) DNA–DNA hybridization determined in microwells using covalent attachment of DNA. Int J Syst Bacteriol 50:1095–1102
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycan based on 2,4-diaminobutyric acid. J Appl Bacteriol 48:459–470
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution well as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20:406–416
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov., an amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715
Huang QY, Dong CZ, Dong RM, Jiang HC, Wang S, Wang GH, Fang B, Ding XX, Niu L, Li X, Zhang CL, Dong HL (2011) Archaeal and bacterial diversity in hot springs on the Tibetan Plateau, China. Extremophiles 15:549–563
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kovacs N (1956) Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 178:703–704
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J Lip Chromatogr 5:2359–2367
Kumar NR, Nair S, Langer S, Busse HJ, Kämpfer P (2008) Altererythrobacter indicus sp. nov., isolated from wild rice (Porteresia coarctata Tateoka). Int J Syst Evol Microbiol 58:839–844
Kwon KK, Woo JH, Yang SH, Kang JH, Kang SG, Kim SJ, Sato T, Kato C et al (2007) Altererythrobacter epoxidivorans gen. nov. sp. nov. an epoxide hydrolase-active, mesophilic marine bacterium isolated from cold-seep sediment, and reclassification of Erythrobacter luteolus Yoon, 2005 as Altererythrobacter luteolus comb. nov. Int J Syst Evol Microbiol 57:2207–2211
Li WJ, Xu P, Schuman P, Zhang YQ, Pukall R, Xu LH, Stackebrandt E, Jiang CL (2007) Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China) and emended description of the genus Georgenia. Int J Syst Evol Microbiol 57:1424–1428
Liang XY, Lin HN, Wang KL, Liao YJ, Lai QL, Xu Y, Wang CY (2016) Altererythrobacter salegens sp. nov., a slightly halophilic bacterium isolated from surface sediment. Int J Syst Evol Microbiol. doi:10.1099/ijsem.0.001708
Locci R (1989) Streptomyces and related genera. In: Williams ST, Sharpe ME, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 4. Williams & Wilkins, Baltimore, pp 2451–2508
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95
Nedashkovskaya OI, Cho SH, Joung Y, Joh K, Kim MN, Shin KS, Oh HW, Bae KS, Mikhailov VV, Kim SB (2013) Altererythrobacter troitsensis sp. nov., isolated from the sea urchin Strongylocentrotus intermedius. Int J Syst Evol Microbiol 63:93–97
Park SC, Baik KS, Choe HN, Lim CH, Kim HJ, Ka JO, Seong CN (2011) Altererythrobacter namhicola sp. nov. and Altererythrobacter aestuarii sp. nov., isolated from seawater. Int J Syst Evol Microbiol 61:709–715
Rainey FA, Silva J, Nobre MF, Silva MT, Costa MS (2003) Porphyrobacter cryptus sp. nov., a novel slightly thermophilic, aerobic, bacteriochlorophyll a-containing species. Int J Syst Evol Microbiol 53:35–41
Reasoner DJ, Geldreich EE (1985) A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (2001) Identification of bacteria by gas chromatography of cellular fatty acids. http://www.microbialid.com/PDF/TechNote_101.pdf
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Song ZQ, Zhi XY, Li WJ, Jiang HC, Zhang CL, Dong HL (2009) Actinobacterial diversity in hot springs in Tengchong (China), Kamachatka (Russia), Nevada (USA). J Gen Microbiol 26:256–263
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882
Waksman SA (1967) The Actinomycetes: a summary of current knowledge. Ronald Press, New York
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Williams ST, Goodfellow M, Alderson G (1989) Genus Streptomyces Waksman and Henrici 1943, 339AL. In: Williams ST, Sharpe ME, Holt JG (eds) Bergey’s Manual of Systematic Bacteriology, vol. 4. Williams & Wilkins, Baltimore, pp 2453–2492
Wu YH, Xu L, Meng FX, Zhang DS, Wang CS, Oren A, Xu XW (2014) Altererythrobacter atlanticus sp. nov., isolated from deep-sea sediment. Int J Syst Evol Microbiol 64:116–121
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH, Xu H, Jiang CL (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family Oxalobacteraceae isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Xue XQ, Zhang KD, Cai F, Dai J, Wang Y, Rahman E, Peng F, Fang CX (2012) Altererythrobacter xinjiangensis sp. nov., isolated from desert sand, and emended description of the genus Altererythrobacter. Int J Syst Evol Microbiol 62:28–32
Xue H, Piao CG, Guo MW, Wang LF, Fang W, Li Y (2016) Description of Altererythrobacter aerius sp. nov., isolated from air, and emended description of the genus Altererythrobacter. Int J Syst Evol Microbiol 66:4543–4548
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2016) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. doi:10.1099/ijsem.0.001755
Zhang GY, Yang YL, Wang LN (2016a) Altererythrobacter aurantiacus sp. nov., isolated from deep-sea sediment. Antonie van Leeuwenhoek 109:1245–1251
Zhang W, Yuan X, Feng QQ, Zhang RG, Zhao XM (2016b) Lv J (2016b), Altererythrobacter buctense sp. nov., isolated from mudstone core. Antonie van Leeuwenhoek 109:793–799
Acknowledgements
We are grateful to Prof. Yu-Guang Zhou (CGMCC, China) for providing the reference type strain and Prof. Aharon Oren, the Hebrew University of Jerusalem, for suggesting the Latin names of the new strains. This work was supported by the Key Project of International Cooperation of Ministry of Science & Technology (MOST) (No. 2013DFA31980), Science and technology infrastructure work project (No. 2015FY110100) and project of China Tobacco Yunnan Industrial Co. Ltd. (No. 2015CP01). W.-J. Li was also supported by Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (2014).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yuan, CG., Chen, X., Jiang, Z. et al. Altererythrobacter lauratis sp. nov. and Altererythrobacter palmitatis sp. nov., isolated from a Tibetan hot spring. Antonie van Leeuwenhoek 110, 1077–1086 (2017). https://doi.org/10.1007/s10482-017-0882-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0882-y