Abstract
A yellow-pigmented, Gram-negative, non-flagellated, rod-shaped bacterial strain, designated M0322T, was isolated from a mudstone core sample of the Mohe Basin, China. Growth of strain M0322T was observed at 15–40 °C (optimum, 30 °C), at pH 5.0–10.0, (optimum, pH 6.0–7.0) and in the presence 0–4 % NaCl (optimum, 0 %). Phylogenetic analyses of 16S rRNA gene sequences revealed that strain M0322T formed a distinct phyletic lineage with the members of the genus Altererythrobacter and is closely related to Altererythrobacter aestuarii JCM 16339T (96.1 %) and Altererythrobacter namhicola JCM 16345T (95.7 %). The only isoprenoid quinone was identified as ubiquinone 10 (Q-10), major polar lipids were determined to be phosphatidylethanolamine, phosphatidylglycerol, phosphatidylcholine, one unidentified glycolipid and three unidentified phospholipids, while major cellular fatty acids were summed feature 8 (C18:1 ω6c and/or C18:1 ω7c), summed feature 3 (C16:1 ω6c and/or C16:1 ω7c) and 11-Methyl C18:1 ω7c. The DNA G+C content of strain M0322T was determined to be 64.6 mol%. Based on the results of the polyphasic taxonomic study, strain M0322T is considered to represent a novel species of the genus Altererythrobacter, for which the name Altererythrobacter buctense sp. nov. is proposed. The type strain is M0322T (=CGMCC 1.12871T = JCM 30112T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Altererythrobacter, a member of the family Erythrobacteraceae (Lee et al. 2005), was first proposed by Kwon et al. (2007). Currently, the genus includes 16 species (http://www.bacterio.net/altererythrobacter.html). The members of the genus Altererythrobacter are characterised as yellow-pigmented, Gram-negative and rod-shaped bacteria, which are lacking of BChl a as a photosynthetic pigment; C18:1 ω7c is the major fatty acid and Q-10 is the main respiratory quinone (Kwon et al. 2007). In this work, a yellow-pigmented strain M0322T, which was isolated from a subterranean sediment sample of Mohe basin permafrost in northeast of China, was subjected to a polyphasic taxonomic investigation in order to establish its taxonomic status,
Materials and methods
Isolation of the bacterial strain
In 2011, 123 drilling core samples were collected from the Mohe basin scientific well MK-2 (53°28′37.80″N, 122°19′23.40″E). Well MK-2 is located in Mohe basin permafrost in northeast of China. With exogenous microbial contamination eliminated by fluorescent microspheres (Colwell et al. 1992), a mudstone core sample (depth, 60.0 m) was separated and ground to powder under liquid nitrogen (SPEX Sample Prep 6870, USA) and sterile conditions (Huang et al. 2014). A portion of the core sample powder (1 g) was serially diluted in a saline solution (0.85 %, w/v), spread on R2A agar (Difco, US) and incubated for one week at 25 °C aerobically. A yellow-pigmented strain M0322T was isolated from the mudstone core sample. The isolate was routinely grown under the same conditions and preserved in R2A broth (without agar) containing glycerol (15 %, v/v) at −80 °C.
Morphological and physiological characteristics
Cell morphology and presence of flagella were observed by the phase-contrast microscopy (Leica DMLS, Germany), transmission (JEM-2100; JEOL) and scanning (SU1510; HITACHI) electron microscopy with cells grown on R2A agar for 3 days at 30 °C. Gliding motility was determined as described by Bernardet et al. (2002). The Gram reaction was performed with 2 % (w/v) crystal violet (Gerhardt 1994). The growth condition was examined on MR agar (per liter distilled water: peptone, 2.75 g; yeast extract, 0.75 g; ferric citrate, 0.05 g; NaCl, 9.72 g; MgCl2, 2. 95 g; MgSO4, 1.65 g; CaCl2, 0.90 g; KCl, 0.28 g; casamino acids 0.25 g; glucose, 0.25 g; soluble starch, 0.25 g; Na-pyruvate, 0.15 g; K2HPO4, 0.15 g; NaHCO3, 0.08 g; KBr, 0.04 g; SrCl2, 17.0 mg; boric Acid, 11.0 mg; Na2SiO4, 2 mg; NaF, 1.2 mg; NH4NO3, 0.8 mg; Na2HPO4, 4 mg; agar, 15 g; pH 7.2 ± 0.2 at 25 °C), R2A agar, marine broth agar (MA, Difco, US), trypticase soy broth agar (TSBA, Difco, US), nutrient agar (NA, Difco, US) and Luria–Bertani agar (LB, Difco, US) for 5 days at 30 °C. The optimum temperature for growth was determined in R2A broth in triplicate at 4, 10, 15, 20, 25, 30, 35, 37, 40 and 42 °C for 3 days on a rotary incubator. The pH range for growth was also determined in R2A broth in triplicate at 30 °C for 3 days on a rotary incubator model, conditions. The optimal pH for growth was tested in R2A broth at pH 4.5–10.0 (at intervals of pH 0.5) by using the following buffer system: pH 4.0–5.0, 0.1 M citric acid/0.1 M sodium citrate; pH 6.0–8.0, 0.1 M KH2PO4/0.1 M NaOH; pH 9.0–10.0, 0.1 M NaHCO3/0.1 M Na2CO3; pH 11.0–12.0, 0.05 M Na2HPO4/0.1 M NaOH. Salt tolerance was examined in R2A broth supplemented with 0–7 % (w/v, at intervals of 1 %) at 30 °C for 3 days. The growth turbidity was recorded using a spectrophotometer (O.D.600 nm; UV-2450, Shimadzu). Anaerobic growth was assessed on R2A agar after incubation for up to 5 day in an anaerobic chamber (CO2/H2/N2, 5:5:90; YQX-II CIMO). Bacteriochlorophyll (BChl) a was detected by using Cary 100 Bio UV–visible Spectrophotometer (Varian, USA), according to the method described by Gich and Overmann (2006). The oxidase and catalase activities were tested using 1 % tetramethyl-p-phenylenediamine (oxidase test strips; bioMérieux) and 3 % (v/v) H2O2, respectively. H2S production was assessed using the methods of Barrow and Feltham (1993). Tests for hydrolysis of starch, casein, gelatin, urea, chitin, Tween 20, and Tween 80 were assessed as described elsewhere (Yuan et al. 2014). Additional physiological and biochemical analyses, including the enzyme activities and carbon source assimilation, were performed in parallel with A. aestuarii JCM 16339T and A. namhicola JCM 16345T by using the API 20 NE, API 20E, API ZYM and API 50CH kits (bioMérieux) according to the instructions of manufacturer, except that the all strips were incubated at 30 °C.
Phylogenetic analysis by the 16S rRNA sequencing and DNA base content
To determine 16S rRNA gene sequences, bacterial DNA was extracted and purified using a bacterial DNA kit (OMEGA, USA). The 16S rRNA gene was amplified by PCR using primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-AAGGAGGTGATCCAGCC-3′). The PCR product was purified using a PCR purification kit (OMEGA) and then ligated into the pMD18-T vector (TaKaRa). The ligation product was transformed into Escherichia coli strain DH5α. The cloned 16S rRNA gene sequence was then sequenced on an ABI 3730XL 96-capillary DNA analyser (Applied Biosystems). The nearly complete 16S rRNA gene sequence (1449 bases) of strain M0322T was determined. The search for phylogenetic neighbours based on 16S rRNA gene sequence similarity was performed using the EzTaxon server (http://www.eztaxon.org/; Kim et al. 2012). Alignment was performed using ARB release 6.0.2 (Ludwig et al. 2004) based on the complete SSU rRNA ‘All-Species Living Tree’ Project (LTP) database (Yarza et al. 2008) release 121 (July 2015). Sequences not included in the LTP database were imported into the LTP database, automatically aligned with the Fast Aligner tool. The alignment of sequences was controlled manually based on secondary structure prediction information. Evolutionary distances were calculated using the algorithm of Kimura’s two-parameter model (Kimura 1980). Distances (distance options according to Kimura’s two-parameter model) and clustering with the maximum-likelihood (Kishino and Hasegawa 1989), neighbour-joining (Saitou and Nei 1987) and maximum-parsimony (Fitch 1971) methods in MEGA release 6.06 (Tamura et al. 2011) were determined by using bootstrap values based on 1000 replications (Felsenstein 1985). The DNA G+C content of strain M0322T was determined as described previously (Zhang et al. 2014).
Analyses of biochemical and chemotaxonomic characteristics
For analyses of biochemical and chemotaxonomic characteristics, A. aestuarii JCM 16339T and A. namhicola JCM 16345T were selected as reference strains. Strain M0322T, A. aestuarii JCM 16339T and A. namhicola JCM 16345T did not grow well on R2A agar and MA medium (Table 1), so a modified MA medium (MR medium) was used as basal medium to grow strain M0322T. Fatty acid analysis was performed as described by Zhang et al. (2014). Bacterial strains were grown on MR at 30 °C at late-exponential phase. Fatty acids were saponified, methylated and extracted from bacterial cells using the standard protocol (MIDI, Sherlock Microbial Identification System, version 6.0). The fatty acids were analyzed by using GC-spell out (6890N; Hewlett Packard) and identified using the TSBA6 database of the Microbial Identification System (Sasser 1990). Polar lipids and isoprenoid quinones were integrally extracted from 100 mg of freeze-dried cell according to the methods described by Minnikin et al. (1984), except that cells were grown on MR agar at 30 °C for 3 days. The polar lipids and isoprenoid quinone were determined as described elsewhere (Yuan et al. 2014).
Results and discussion
Morphological and physiological characteristics
Strain M0322T was observed to be Gram-stain negative, facultative anerobic, non-motile, non-flagellated short rods. The scanning electron micrograph of strain M0322T is included in Supplementary Fig. S1. It was found that strain M0322T does not contain BChl a as a photosynthetic pigment, is positive for β-glucosidase, aesculin hydrolysis and β-galactosidase, but negative for reduction of nitrate to nitrite, indole production, d-glucose fermentation, arginine dihydrolase, urease activity, gelatin hydrolysis and utilization of d-glucose, l-arabinose, d-mannose, d-mannitol, N-acetylglucosamine, maltose, potassium gluconate, capric acid, adipic acid, malic acid, trisodium citrate and phenylacetic acid. According to API 20E, strain M0322T was found to be positive for β-galactosidase and acetoin production, negative for lysine decarboxylase, ornithine decarboxylase, citrate utilization, H2S production, urease activity, tryptophan deaminase activity, indole production, gelatinase and acid production from glucose, mannitol, inositol, sorbitol, rhamnose, sucrose melibiose, amygdalin and L-arabinose. According to API 50CH, strain M0322T was found to be positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, β-glucuronidase, α-glucosidase and β-glucosidase; weakly positive for valine arylamidase; negative for lipase (C14), α-galactosidase, N-acetyl-β-glucosaminidase, α-mannosidase and β-fucosidase (API ZYM). Acid is only produced from aesculin ferric citrate (API 50CH). Other morphological, physiological and biochemical characteristics of strain M0322T are given in the species description. The differential morphological, physiological, and biochemical characteristics between strain M0322T and reference strains are shown in Table 1.
Chemotaxonomic characteristics
The whole-cell fatty acid compositions of strain M0322T and the reference strains are shown in Table 2. The major fatty acids (>10 % of the total fatty acids) of strain M0322T were determined to be summed feature 3 (29.5 %, C16:1 ω6c and/or C16:1 ω7c), summed feature 8 (27.8 %, C18:1 ω6c and/or C18:1 ω7c) and 11-Methyl C18:1 ω7c (22.0 %). Strain M0322T was found to possess summed feature 3 and summed feature 8 as the major fatty acids, which are similar to those detected in A. aestuarii JCM 16339T and A. namhicola JCM 16345T. However, 11-Methyl C18:1 ω7c was found to be abundant in strain M0322T, while the proportion of C17:1 ω6c is higher in A. namhicola JCM 16345T. The only isoprenoid quinone of strain M0322T was determined to be ubiquinone 10 (Q-10), which is the characteristic ubiquinone for the genus Altererythrobacter. The polar lipid profile of M0322T was found to consist of phosphatidylethanolamine, phosphatidylglycerol, phosphatidylcholine, one unidentified glycolipid and three unidentified phospholipids (Fig. S3). The polar lipid profile of strain M0322T was found to be similar to that of A. aestuarii JCM 16339T, but different to that of A. namhicola JCM 16345T (Fig. S3).
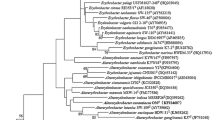
Phylogenetic analysis of 16S rRNA sequences
The 16S rRNA gene sequence analysis of the almost complete 16S rRNA gene sequence (1449 bp) of strain M0322T indicated that strain M0322T formed a weak cluster (below 50 % of bootstrap support) with two species of the genus Altererythrobacter, A. aestuarii JCM 16339T (96.1 %) and A. namhicola JCM 16345T (95.7 %). Analysis of the maximum-likelihood tree (Fig. 1) and neighbor-joining tree (Fig. S2) showed that the genus Altererythrobacter is not monophyletic. The species of the genus Altererythrobacter are grouped with bacteria of the genera Croceicoccus, Sphingopyxis and Novosphingobium and with bacteria belonging to the family Sphingomonadaceae (the genera Novosphingobium and Sphingopyxis). However, the characteristics of the novel isolate are consistent with the description of the genus Altererythrobacter with regard to morphological, biochemical and chemotaxonomic properties supporting the affiliation of strain M0322T with the genus Altererythrobacter. At the same time on the basis of phylogenetic distances below 97 % between strain M0322T and two phylogenetically closest species, A. aestuarii JCM 16339T and A. namhicola JCM 16345T, it can be concluded that strain M0322T represents a distinct species of the genus Altererythrobacter. Furthermore, strain M0322T can be readily distinguished from other Altererythrobacter species by the combination of many of the physiological and biochemical features (Table 1). Therefore, strain M0322T is considered to represent a novel species within the genus Altererythrobacter, for which the name Altererythrobacter buctense sp. nov. is proposed.
A maximum-likelihood (ML) phylogenetic tree based on the 16S rRNA gene sequences of strain M0322T and representatives of related taxa. Numbers at the nodes indicate the bootstrap values (greater than 50 %) expressed as a percentage of 1000 replicates. Bar represents 0.02 substitutions per nucleotide position
Description of Altererythrobacter buctense sp. nov
Altererythrobacter buctense (buc.ten’se. N.L. masc. adj. Referring to the acronym BUCT, Beijing University of Chemical Technology, where the strain was identified).
Cells are Gram-negative, facultative anerobic, non-motile, non-flagellated, short rods (approximately 0.2–0.4 × 0.5–0.9 μm). Colonies on R2A are yellow-pigmented, smooth, circular and convex. Grows at 15–40 °C (optimum, 30 °C), pH 5.0–10.0 (optimum, pH 6.0–7.0) and in the presence of 0–4.0 % NaCl (optimum, 0 %). Does not contain BChl a as a photosynthetic pigment. Methanol-soluble pigment is characterised by absorption maxima at 449 and 478 nm. Grows well on MR agar, R2A agar, LB and TSBA, but poorly on MA and does not grow on NA. Positive for hydrolysis of Tween 20 and urea, oxidase and catalase activities. Negative for hydrolysis of starch, tyrosine, gelatin, cellulose, chitin and Tween 80. H2S is not produced. The only isoprenoid quinone is identified to be Q-10. The major polar lipids are phosphatidylethanolamine, phosphatidylglycerol, phosphatidylcholine, one unidentified glycolipid and three unidentified phospholipids. The major fatty acids (>10 %) are Summed Feature 3 (C16:1 ω6c and/or C16:1 ω7c), Summed Feature 8 (C18:1 ω6c and/or C18:1 ω7c) and 11-Methyl C18:1 ω7c. The DNA G+C content of the type strain is 64.6 mol%.
The type strain, M0322T (=CGMCC 1.12871T = JCM 30112T), was isolated from the Mohe Basin, China. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain M0322T is KJ599648.
References
Barrow GI, Feltham RKA (eds.) (1993) Cowan and Steelham RKA (eds.) (1993) the experimental work.esearc, 3rd edn. Cambridge University Press, Cambridge
Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Colwell FS, Stormberg GJ, Phelps TJ, Birnbaum SA, McKinley J, Rawson SA, Veverka C, Goodwin S, Long PE, Russell BF, Garland T, Thompson D, Skinner P, Grover S (1992) Innovative techniques for collection of saturated and unsaturated subsurface basalts and sediments for microbiological characterisation. J Microbiol Methods 15:279–292
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Gerhardt P (1994) Methods for general and molecular bacteriology. American Society for Microbiology, Washington DC
Gich F, Overmann J (2006) Sandarakinorhabdus limnophila gen. nov., sp. nov., a novel bacteriochlorophyll a-containing, obligately aerobic bacterium isolated from freshwater lakes. Int J Syst Evol Microbiol 56:847–854
Huang F, Zhang Y, Zhu Y, Wang P, Lu J, Lv J (2014) Flavobacterium qiangtangensis sp. nov., isolated from Qiangtang basin in Qinghai-Tibetan plateau, China. Curr Microbiol 69:234–239
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kishino H, Hasegawa M (1989) Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol 29:170–179
Kwon KK, Woo JH, Yang SH, Kang JH, Kang SG, Kim SJ, Sato T, Kato C et al (2007) Altererythrobacter epoxidivorans gen. nov., sp. nov., an epoxide hydrolase-active, mesophilic marine bacterium isolated from cold-seep sediment, and reclassification of Erythrobacter luteolus Yoon et al. 2005 as Altererythrobacter luteolus comb. nov. Int J Syst Evol Microbiol 57:2207–2211
Lee KB, Liu CT, Anzai Y, Kim H, Aono T, Oyaizu H (2005) The hierarchical system of the ‘Alphaproteobacteria’: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int J Syst Evol Microbiol 55:1907–1919
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S et al (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Park SC, Baik KS, Choe HN, Lim CH, Kim HJ, Ka JO, Seong CN (2011) Altererythrobacter namhicola sp. nov. and Altererythrobacter aestuarii sp. nov., isolated from seawater. Int J Syst Evol Microbiol 61:709–715
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI Inc, Newark
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimonymethods. Mol Biol Evol 28:2731–2739
Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH, Ludwig W, Gllecular E, Rosselló-Móra R (2008) The all-species living tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 31:241–250
Yuan X, Nogi Y, Tan X, Zhang RG, Lv J (2014) Arenimonas maotaiensis sp. nov., isolated from fresh water. Int J Syst Evol Microbiol 64:3994–4000
Zhang RG, Tan X, Zhao XM, Deng J, Lv J (2014) Moheibacter sediminis gen. nov., sp. nov., a novel member of the family Flavobacteriaceae, isolated from the sediment, and emended descriptions of Empedobacter brevis, Wautersiella falsenii and Weeksella virosa. Int J Syst Evol Microbiol 64:1481–1487
Acknowledgments
This work was supported by grants from the National Special Research Fund of China (Gas Hydrate Resource Exploration and Production Testing Project. Grant No. GZHL20110317). We thank L. Han, L. Jia and X.M. Feng for their assistance in the experimental work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, W., Yuan, X., Feng, Q. et al. Altererythrobacter buctense sp. nov., isolated from mudstone core. Antonie van Leeuwenhoek 109, 793–799 (2016). https://doi.org/10.1007/s10482-016-0679-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0679-4