Abstract
Computational drug repositioning has been proven as a promising and efficient strategy for discovering new uses from existing drugs. To achieve this goal, a number of computational methods have been proposed, which are based on different data sources of drugs and diseases. These methods approach the problem using either machine learning- or network-based models with an assumption that similar drugs can be used for similar diseases to identify new indications of drugs. Therefore, similarities between drugs and between diseases are usually used as inputs. In addition, known drug-disease associations are also needed for the methods as prior information. It should be noted that those associations are still not well established due to the fact that many of marketed drugs have been withdrawn and this could affect the outcome of the methods. In this study, we propose a novel method named RLSDR (Regularized Least Square for Drug Repositioning) to find new uses of drugs. More specifically, it relies on a semi-supervised learning model, Regularized Least Square, thus it does not require definition of non-drug-disease associations as previously proposed machine learning-based methods. In addition, the similarity between drugs measured by chemical structures of drug compounds and the similarity between diseases which share phenotypes can be represented in a form of either similarity network or similarity matrix as inputs of the method. Moreover, instead of using a gold-standard set of known drug-disease associations, we construct an artificial set of the associations based on known disease-gene and drug-target associations. Experiment results demonstrate that RLSDR achieves better prediction performance on the artificial set of drug-disease associations than that on the gold-standard ones in terms of area under the Receiver Operating Characteristic (ROC) curve (AUC). In addition, it outperforms two representative network-based methods irrespective of the prior information of drug-disease associations. Novel indications for a number of drugs are also identified and validated by evidences from a different data resource.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
De novo drug discovery is a costly and time-consuming task due to failures spreading throughout the drug development pipeline. Indeed, it takes billions of dollars of investment and an average of 9–12 years to bring a new drug to the market (Dickson and Gagnon 2009; Ashburn and Thor 2004; Hughes et al. 2011). Therefore, drug repositioning is an emerging approach to detect new clinical indications for existing drugs, or for those that are in the development pipeline (Ashburn and Thor 2004; Hurle et al. 2013). For example, the monoclonal antibody bevacizumab, originally developed for metastatic colon cancer and non-small cell lung cancer by inhibiting angiogenesis, is now being used to slow or reverse abnormal vascularization of the retina in exudative macular degeneration (Rich et al. 2006). In addition, it was reported the new uses of sildenafil for erectile dysfunction and pulmonary hypertension, thalidomide for severe erythema nodosum leprosum, and retinoic acid for acute promyelocytic leukemia (Aronson 2007; Sirota et al. 2011). Other examples of drug repositioning can be found in the study of Ashburn and Thor (2004) and Sardana et al. (2011). However, most of the successful examples are from clinical observations. In addition, the underlying molecular mechanisms are often not clear for many cases. Therefore, computational methods are developed to effectively find new uses of drugs against various diseases and to understand the underlying mechanisms.
Two main approaches have been proposed for drug repositioning including: (i) machine learning-based and (ii) network-based. It was shown that integration of data of drugs and diseases has been proven to be an effective method in drug repositioning (Zhang et al. 2014). Therefore, both of the approaches usually integrate various drug- and disease-related data in machine learning/network-based models to infer new drug-disease associations. For instance, Gottlieb et al. (2011) integrated various disease-related features including phenotypic and genetic features. Then, a logistic regression classifier is used to predict novel drug indications. Rather than disease-related features, drug-related features built from drug chemical structure similarity, drug molecular target similarity and drug gene expression similarity, were used to predict drug therapeutic class (Napolitano et al. 2013). These features were then merged into a single drug similarity matrix and used as a kernel for Support Vector Machines classifier. In addition, Menden et al. developed the machine learning-based models, a feed-forward perceptron neural network and a random forest regression, to predict the response of cancer cell lines to drug treatment using cancer genomic features of the cell lines and chemical properties of drugs (Menden et al. 2013). Instead of feature-based data integration as in those studies, Wang et al. (2013) integrated molecular structure, molecular activity, and phenotype data using kernel-based data integration via a multiple kernel learning technique. A common limitation of those machine learning-based methods is that the definition of non-drug-disease associations are considered as a negative training set used in binary classification models. This is because of the fact in biomedicine that the not yet observed association does not mean that the association does not exist.
Network-based analysis is another widely used strategy for computational drug repositioning (Wu et al. 2013). Based on the same assumption that similar drugs can be used to treat similar diseases, network-based methods also rely on similarity between drugs and diseases. However, the similarities are presented in similarity networks rather than similarity matrices (i.e., kernel matrices). In addition, prior information such as known drug-target interactions, known disease-gene associations and known drug indications are also integrated. For example, a causal network was recently built (Li and Lu 2013), which is a multilayered pathway of gene, disease and drug target, to identify new therapeutic uses of existing drugs. In the causal network, the transition likelihood of each chain was estimated on the basis of known drug-disease treatment association. However, most studies first built a heterogeneous network including a drug similarity network and a disease similarity network, which are connected to each other by known drug-disease associations, and then a network-based algorithm was used to infer novel associations between drugs and diseases. For instance, Wu et al. applied network clustering to a drug-disease heterogeneous network to identify closely connected modules of disease and drugs, which can be used for extracting possible drug-disease pairs for drug repositioning (Wu et al. 2013). A random walk with restart (RWRH) model was used to infer new indications for approved drugs on drug-disease heterogeneous networks (Liu et al. 2016). In addition, other heterogeneous graph-based inference (HGBI) methods, which were successfully used for prediction of drug-target interactions (Wang et al. 2013), can be used for drug repositioning.
It is obvious that the prediction performances of both machine learning- and network-based methods are dependent on similarity matrices/networks. Besides, prior information represented in known drug-disease associations in a form of an adjacency matrix/a bipartite network for machine learning- and network-based, respectively, also affect the prediction performance. However, those associations are still not well established due to the fact that many of marketed drugs have been withdrawn. In this study, we propose to use a novel method named RLSDR, which rely on a semi-supervised learning method, i.e., Regularized Least Square, to overcome a limitation of supervised classification methods since it does not require to clearly define the negative training set. In addition, it can make the use of the heterogeneous network of drugs and diseases as input matrices. Indeed, drug and disease similarity networks can be represented as similarity matrices and a bipartite network of known drug-disease associations can be used as a manner of an adjacency matrix. Due to the limitation of available known drug-disease associations as gold-standard sets, instead of using these associations, we built an artificial set of the associations based on the shared proteins which are targets of a drug and known to be associated with a disease. This approach is based on an assumption that the shared proteins can be deemed as the molecular origin of diseases and drugs development, which are taking part in common biological processes perturbed under disease state or under drug administration. Therefore, these shared proteins may imply potential drug repositioning.
To assess the overall prediction performance of RLSDR, we first compared itself when using the constructed artificial set and a gold-standard set collected from Gottlieb et al. (2011) of drug-disease associations. The experiment result demonstrated that RLSDR performed better on the artificial set. After that, we compared RLSDR with two representative network-based methods, one working solely on the disease similarity network and one applied to the heterogeneous network of drugs and diseases. Experiment results also indicated that RLSDR outperformed both of them. To assess the prediction ability of novel drug-disease associations of RLSDR, we used the gold-standard (which was collected from Gottlieb et al. (2011)) set as training set and another gold-standard dataset of drug-disease associations from Martínez et al. (2015) for the validation. As a result, a total of 19 out of 75 drugs, which are common between the two gold-standard databases, has found as novel indications.
2 Materials and Methods
2.1 Drug Similarity
It is generally accepted that drugs with similar chemical structures would have similar therapeutic functions and can be used to treat similar diseases. Therefore, we calculate similarity between 7838 drugs from KEGG database (Kanehisa et al. 2009) using SIMCOMP tool (Hattori et al. 2010), which calculates the similarity between drugs based on their chemical structures. More specifically, SIMCOMP has been originally developed as a graph-based method for comparing chemical structures, which searches for the maximal cliques in the association graph as the maximum common induced subgraph. However, the current version of SIMCOMP can also compute the maximum common edge subgraph, which is faster because of the small number of nodes in an association graph. The calculation results in 887,883 interactions having positive similarity between every pair of the drugs. This drug similarity network can be represented as a similarity matrix \(W_{Dr}\), where an element \(\left( {W_{Dr} } \right)_{i,j}\) represents similarity between two drugs \(dr_{i}\) and \(dr_{j} .\)
2.2 Disease Similarity
We collect a phenotypic disease similarity matrix (\(W_{D}\)) from MimMiner (van Driel et al. 2006), where an element \(\left( {W_{D} } \right)_{i,j}\) of the matrix represents the degree of similarity between two disease phenotypes \(d_{i}\) and \(d_{j}\). The disease phenotypes are OMIM records (Amberger et al. 2009). More specifically, the full-text and clinical synopsis fields of more than 5000 records that describe genetic disorders were used. Then, a feature vector was built for each OMIM record based on the anatomy and the disease sections of the medical subject headings vocabulary (MeSH) (Lipscomb 2000), where MeSH concepts serve as phenotype features characterizing OMIM records. Finally, the similarity between two disease phenotypes \(d_{i}\) and \(d_{j}\) is calculated by cosine of angles of the two corresponding feature vectors. By selecting only k neighbors which have largest similarities for each node, a phenotypic disease similarity network can be constructed.
2.3 Gold-Standard Sets of Known Drug-Disease Associations
Known drug-disease associations assembled by Gottlieb et al. (2011) are considered as a first gold-standard. This contains 1933 associations between 593 drugs collected from DrugBank (Law et al. 2014) and 313 diseases. After mapping DrugBank identifier of these drugs to KEGG (Kanehisa et al. 2009) identifier, and disease name with title of disease phenotype in OMIM (Amberger et al. 2009), we finally have 1297 associations between 403 drugs and 257 diseases. This first gold-standard is called as PREDICT gold-standard since this is the name of the method proposed in Gottlieb et al. (2011). The second gold-standard of drug-disease associations, DNdataset, was collected from Martínez et al. (2015). This contains 1008 drug-disease associations between 1490 drugs from DrugBank and 4517 diseases from disease ontology (DO) (Kibbe et al. 2015). After mapping with KEGG identifier and disease phenotype name from OMIM, we finally obtained 231 associations between 149 drugs and 76 diseases.
2.4 Construction of Artificial Drug-Disease Associations
Due to the limitation in available gold-standard drug-disease associations, we here define an association between a drug and a disease using shared genes/gene products. This definition is derived from underlying mechanisms of drugs and diseases that the common biological processes perturbed under disease state by associated genes or under drug administration by binding mediated targets may imply potential drug repositioning. To construct these associations, we first collect known drug-target interactions between 1198 drugs and 1421 targets from KEGG (Kanehisa et al. 2009), and known disease-gene associations between 21,249 diseases and 15,448 genes from OMIM (Amberger et al. 2009). An association between a drug dr and a disease d is defined if there is significant overlap between the known target set of dr and the known associated gene set of d using hypergeometric distribution
where
-
N is number of targets in KEGG database
-
n is number of known target of drug dr
-
K is number of gene known to be associated with disease d
-
k is number of shared target/gene between drug dr and disease d.
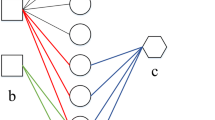
By selecting only drug-disease pair having p ≤ 0.05, we finally have 6452 significant associations between 785 drugs and 1181 diseases. We denote these associations as an artificial set. Figure 1a illustrates the construction of the artificial set of drug-disease associations.
Illustration of RLSDR method for drug repositioning. a Construction of artificial drug-disease associations. Known drug-target interactions and known disease-gene associations are collected from KEGG and OMIM, respectively. Then, an association between a drug and a disease is determined if there is a significant overlap between known target set of the drug and known associated gene set of the disease. b RLSDR is used to rank candidate diseases to find new uses of a drug dr1 in a framework of a heterogeneous network of drugs and diseases
2.5 Regularized Least Square for Drug Repositioning (RLSDR)
RLSDR is proposed based on Regularized Least Square (RLS) learning technique for inferring novel drug-disease associations. This is a semi-supervised learning and global method since it can rank drug-disease associations for all the drugs simultaneously without the negative samples. This method was designed to construct a continuous classification function which can determine the association probability between each drug and a given disease (i.e., the higher this probability is, the more each drug is related to a given disease). To this end, a RLS classifier was constructed by defining and minimizing a cost function. This cost function was trained in the drug similarity network and the disease similarity network, and then it was used to optimize the classification function (Chen and Yan 2014). Formally, the optimal classifier in these two spaces was defined as follows:
where
-
\(F_{Dr}^{*}\) and \(F_{D}^{*}\) are optimal classification functions in the drug and disease spaces, respectively as follows:
$$F_{Dr}^{*} = W_{Dr} \left( {W_{Dr} + \eta_{Dr} I_{Dr} } \right)A^{T}$$$$F_{D}^{*} = W_{D} \left( {W_{D} + \eta_{D} I_{D} } \right)A$$ -
w is the weight between these two spaces. \(\eta_{Dr}\) and \(\eta_{D}\) are trade-off parameters in the drug and disease spaces, respectively.
-
\(I_{Dr}\) and \(I_{D}\) are identity matrices with the same size as matrices \(W_{Dr}\) and \(W_{D}\), respectively.
-
\(A\left( {mn} \right)\) is an association matrix, where (A)i,j = 1 if drug dri is associated with disease dj, otherwise (A)i,j = 0. m and n are the number of drugs and diseases in the drug similarity networks and the disease similarity network, respectively.
Figure 1b demonstrates the integration of drug and disease similarity networks and drug-disease associations into a heterogeneous network and how RLSDR ranks diseases to find potential candidates to be repositioned for a given drug.
2.6 Performance Evaluation
Ranking performance was assessed through the leave-one-out cross-validation (Shortly called LOOCV) process for each drug, and then the overall prediction performance of a method is an averaged value over all drugs in a set of known drug-disease associations. Given a drug dr, a set of known dr-associated diseases (S) and a set of candidate diseases (C) are given. A disease s∈D was held out for validation and the remaining known drug-associated diseases were specified to a set of source nodes (i.e., S = D\{s}) for RWR. For RLSDR and HGBI, (A)dr,s is set to 0. The network-based ranking algorithms were used to prioritize all the candidate diseases. This process was repeated by setting every s∈D to a held-out disease. For a reliable performance comparison, we drew the receiver operating characteristic (ROC) curves and computed the area under the curve (AUC) value based on the rank of held-out disease s and diseases in set C∪{s}. More specifically, given a thresholdτ, we counted TP (true positives), FN (false negatives), FP (false positives), and TN (true negatives), which were formally defined as follows:
where rank(s), rank(c) and I(∙) denote the rank of s, the rank of a candidate disease c out of the set C∪{s} and the indicator function, respectively. Then, we defined sensitivity and (1-specificity) as follows:
By varying τ from one to the number of diseases in the set C∪{s}, the relationship between sensitivity and (1-specificity) was plotted. The ROC curve is the curve constructed based on those pairs of values, and the AUC is the area under the ROC curve. In this study, we considered candidate disease set as all diseases that are not known to be associated with the drug dr.
3 Results and Discussion
Prediction performance was assessed through the leave-one-out cross-validation (Shortly called LOOCV) process for each drug, and then the overall prediction performance of a method is an averaged value over all drugs in a set of either artificial or gold-standard drug-disease associations.
3.1 Prediction Performance of RLSDR on Artificial and Gold-Standard Sets of Drug-Disease Associations
In this section, we show how well RLSDR method can predict novel drug-disease associations using artificial drug-target association data. More specifically, we compare the prediction performance of RLSDR using the artificial and PREDICT gold-standard sets of drug-disease associations. To this end, we set \(\eta_{Dr} = \eta_{D} = 1\) and vary the weight parameter in (0, 1) then calculate the prediction performance in terms of AUC value for each drug in each association set. Then, the final performance is averaged over all AUC values. Figure 2 shows that RLSDR on the artificial set is better than that on the gold-standard set for all weight parameter (w) varied in (0, 1). This figure also shows stable performance while changing weight w. In summary, the result indicates that artificial drug-disease associations help improve the inference of new uses of existing drugs.
3.2 Performance Comparison with Existing Methods
In this section, we compare the prediction performance of RLSDR with two network-based methods. First, a random walk with restart (RWR) model was successfully used for prediction of disease-associated genes (Kohler et al. 2008; Le and Kwon 2012; Le and Dang 2016), microRNAs (Le 2015a, b; Le et al. 2017) and protein complexes (Le 2015c) based on a homogeneous network of genes/proteins/miRNAs. We adopted this model to infer novel drug-disease associations using a homogeneous network of diseases (i.e., the disease similarity network, see Sect. 2). In addition, another heterogeneous graph-based inference (HGBI) method, which was successfully used for prediction of drug-target interactions (Wang et al. 2013) and disease-associated miRNAs (Chen et al. 2016), is used for drug repositioning. HGBI is based on the guilt-by-association principle on a heterogeneous network of drugs and diseases and predicts new drug-disease associations by iteratively updating the measure of strength between unlinked drug-disease pairs by taking all the paths in the network into account. Figure 2 demonstrates that the prediction performance of RLSDR is stable while changing weight w, therefore we set w = 0.5 when comparing with other existing methods. Experiment result also shows that HGBI is stable while we vary the decay factor (α) in (0, 1) (data not shown), thus we set α = 0.5 for comparison. For RWR, we also change the back probability in (0, 1) and found that the best performances were achieved with \(\gamma = 0.7\) and \(\gamma = 0.1\) for PREDICT gold-standard and for artificial sets, respectively (data not shown). In addition, due to using LOOCV for assessing prediction performance, only drugs known to be associated with at least two diseases are valid for RWR. Therefore, we also rerun RLSDR for drugs having at least two associated diseases when comparing with RWR. First, these three methods are compared on artificial drug-disease association dataset with the best setting for each method. Figure 3a indicates that RLSDR (AUC = 0.97) is better than that of RWR (AUC = 0.76) and HGBI (AUC = 0.62). Then, we compare these three methods on gold-standard drug-disease association dataset. Similarly, Fig. 3b shows that RLSDR (AUC = 0.93) is superior to both RWR (AUC = 0.79) and HGBI (AUC = 0.52). Taken together, our method achieves better prediction performance than some other existing methods irrespective of drug-disease association datasets. This also indicates that RLSDR, which is based on a semi-supervised learning model, has overcome limitations of two classification supervised learning models and made use effectively the heterogeneous network framework of network-based methods.
3.3 Prediction of Novel Indications for Existing Drugs
In this experiment, we try to predict novel indications of existing drugs. To achieve this goal, we use the PREDICT gold-standard drug-disease association set collected from Gottlieb et al. (2011) (see Sect. 2). Then, we infer novel drug-disease associations for each drug in the set by selecting top 100 ranked diseases for each drug. These diseases could be novel promising indications. To validate these indications, we use another gold-standard dataset of drug-disease associations DNdataset collected from Martínez et al. (Martínez et al. 2015) (see Sect. 2). There are 75 drugs common between the two gold-standard datasets. Table 1 shows novel indications of nineteen drugs out of them have found DNdataset. For example, octreotide (KEGG ID: D00442), which was designed to treat patients with diabetes mellitus, is found its new use in patients with acromegaly. Acromegaly is abnormal growth of the hands, feet, and face, caused by overproduction of growth hormone by the pituitary gland. Similarly, ciclesonide (KEGG ID: D01703) is known to treat asthma, nasal polyps, and aspirin intolerance. However, RLSDR shows that ciclesonide can be used for allergic rhinitis. Finally, meclofenamic acid (KEGG ID: D02341) can be used for osteoarthritis instead of only for patients having insensitivity to pain with hyperplastic myelinopathy as original developed.
4 Conclusions
Computational drug repositioning has been proven as an effective strategy in drug discovery, since it saves both time and cost to get a drug to go to the market. It also helps to find new uses of existing/experimental drugs, which have already passed toxicity validation. Computational methods for drug repositioning fall into two main categories (i.e., machine learning- and network-based approaches), where both of them make use of drug- and disease-related information in a form of similarity matrix/network. In addition, prior information about known drug-disease associations also helps infer new uses of drugs effectively. However, this association set is very limited because of the high cost and time consumption of current drug discovery pipeline. In addition, a number of approved drugs have been withdrawn from the market due to side-effects. Binary classification learning methods proposed for the problem also have limitations in defining negative training set as non-drug-disease associations. Therefore, in this study, we proposed a novel method, RLSDR, which is based on a semi-supervised learning model, thus overcome the limitations of the binary classification-based methods. In addition, we constructed an artificial drug-disease association based on shared target proteins which are both known to be associated to a disease as well as known to be targets of a drug. As a result, RLSDR performed better on the artificial set compared to the gold-standard set collected from a previous study, which then has been used in other studies recently. Moreover, RLSDR also outperforms some other existing methods on both the drug-disease association set. Taken together, RLSDR, which overcomes both limitations of previous machine learning-based methods and the lack of known drug-disease associations, could be an effective strategy for drug repositioning.
References
Amberger J, Bocchini CA, Scott AF, Hamosh A (2009) McKusick’s online mendelian inheritance in man (OMIM®). Nucleic Acids Res 37(suppl 1):D793–D796
Aronson JK (2007) Old drugs—new uses. Br J Clin Pharmacol 64(5):563–565
Ashburn TT, Thor KB (2004) Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 3(8):673–683
Chen X, Yan G-Y (2014) Semi-supervised learning for potential human microRNA-disease associations inference. Sci Rep 4:5501
Chen X, Yan CC, Zhang X, You Z-H, Huang Y-A, Yan G-Y (2016) HGIMDA: heterogeneous graph inference for miRNA-disease association prediction. Oncotarget 7(40):65257–65269
Dickson M, Gagnon JP (2009) The cost of new drug discovery and development. Discov Med 4(22):172–179
Gottlieb A, Stein GY, Ruppin E, Sharan R (2011) PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol 7:496
Hattori M, Tanaka N, Kanehisa M, Goto S (2010) SIMCOMP/SUBCOMP: chemical structure search servers for network analyses. Nucleic Acids Res 38(suppl 2):W652–W656
Hughes JP, Rees S, Kalindjian SB, Philpott KL (2011) Principles of early drug discovery. Br J Pharmacol 162(6):1239–1249
Hurle MR, Yang L, Xie Q, Rajpal DK, Sanseau P, Agarwal P (2013) Computational drug repositioning: from data to therapeutics. Clin Pharmacol Ther 93(4):335–341
Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M (2009) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38(suppl 1):D355–D360
Kibbe WA, Arze C, Felix V, Mitraka E, Bolton E, Fu G, Mungall CJ, Binder JX, Malone J, Vasant D, Parkinson H, Schriml LM (2015) Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res 43(D1):D1071–D1078
Kohler S, Bauer S, Horn D, Robinson P (2008) Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet 82(4):949–958
Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, Tang A, Gabriel G, Ly C, Adamjee S, Dame ZT, Han B, Zhou Y, Wishart DS (2014) DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 42(D1):D1091–D1097
Le D-H (2015a) Disease phenotype similarity improves the prediction of novel disease-associated microRNAs. In: Information and computer science (NICS), 2015 2nd national foundation for science and technology development conference on 16–18 Sept 2015, pp 76–81
Le D-H (2015b) Network-based ranking methods for prediction of novel disease associated microRNAs. Comput Biol Chem 58:139–148
Le D-H (2015c) A novel method for identifying disease associated protein complexes based on functional similarity protein complex networks. Algorithms Mol Biol 10(1):14
Le D-H, Dang V-T (2016) Ontology-based disease similarity network for disease gene prediction. Vietnam J Comput Sci 3:1–9
Le D-H, Kwon Y-K (2012) GPEC: a Cytoscape plug-in for random walk-based gene prioritization and biomedical evidence collection. Comput Biol Chem 37:17–23
Le D-H, Verbeke L, Son LH, Chu D-T, Pham V-H (2017) Random walks on mutual microRNA-target gene interaction network improve the prediction of disease-associated microRNAs. BMC Bioinform 18(1):479
Li J, Lu Z (2013) Pathway-based drug repositioning using causal inference. BMC Bioinform 14(16):S3
Lipscomb CE (2000) Medical subject headings (MeSH). Bull Med Libr Assoc 88(3):265–266
Liu H, Song Y, Guan J, Luo L, Zhuang Z (2016) Inferring new indications for approved drugs via random walk on drug-disease heterogenous networks. BMC Bioinform 17(17):539
Martínez V, Navarro C, Cano C, Fajardo W, Blanco A (2015) DrugNet: network-based drug–disease prioritization by integrating heterogeneous data. Artif Intell Med 63(1):41–49
Menden MP, Iorio F, Garnett M, McDermott U, Benes CH, Ballester PJ, Saez-Rodriguez J (2013) Machine learning prediction of cancer cell sensitivity to drugs based on genomic and chemical properties. PLoS ONE 8(4):e61318
Napolitano F, Zhao Y, Moreira VM, Tagliaferri R, Kere J, D’Amato M, Greco D (2013) Drug repositioning: a machine-learning approach through data integration. J Cheminform 5(1):30
Rich RM, Rosenfeld PJ, Puliafito CA, Dubovy SR, Davis JL, Flynn HWJ, Gonzalez S, Feuer WJ, Lin RC, Lalwani GA, Nguyen JK, Kumar G (2006) SHORT-term safety and efficacy of intravitreal bevacizumab (avastin) for neovascular age-related macular degeneration. Retina 26(5):495–511
Sardana D, Zhu C, Zhang M, Gudivada RC, Yang L, Jegga AG (2011) Drug repositioning for orphan diseases. Brief Bioinform 12:346–356
Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, Sage J, Butte AJ (2011) Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med 3:96ra77
van Driel MA, Bruggeman J, Vriend G, Brunner HG, Leunissen JAM (2006) A text-mining analysis of the human phenome. Eur J Hum Genet 14(5):535–542
Wang Y, Chen S, Deng N, Wang Y (2013a) Drug repositioning by kernel-based integration of molecular structure, molecular activity, and phenotype data. PLoS ONE 8(11):e78518
Wang W, Yang S, Li J (2013b) Drug target predictions based on heterogeneous graph inference. Biocomputing 18:53–64
Wu Z, Wang Y, Chen L (2013a) Network-based drug repositioning. Mol BioSyst 9(6):1268–1281
Wu C, Gudivada RC, Aronow BJ, Jegga AG (2013b) Computational drug repositioning through heterogeneous network clustering. BMC Syst Biol 7(5):S6
Zhang P, Wang F, Hu J (2014) Towards drug repositioning: a unified computational framework for integrating multiple aspects of drug similarity and disease similarity. In: AMIA annual symposium proceedings. American Medical Informatics Association, p 1258
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Le, DH., Nguyen-Ngoc, D. Drug Repositioning by Integrating Known Disease-Gene and Drug-Target Associations in a Semi-supervised Learning Model. Acta Biotheor 66, 315–331 (2018). https://doi.org/10.1007/s10441-018-9325-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10441-018-9325-z