Abstract
We provide an innovative, bioengineering, mechanobiology-based approach to rapidly (2-h) establish the in vivo metastatic likelihood of patient tumor-samples, where results are in direct agreement with clinical histopathology and patient outcomes. Cancer-related mortality is mostly due to local recurrence or to metastatic disease, thus early prediction of tumor-cell-fate may critically affect treatment protocols and survival rates. Metastasis and recurrence risks are currently predicted by lymph-node status, tumor size, histopathology and genetic testing, however, these are not infallible and results may require days/weeks. We have previously observed that subpopulations of invasive cancer-cells will rapidly (1–2 h) push into the surface of physiological-stiffness, synthetic polyacrylamide gels, reaching to cell-scale depths, while normal or noninvasive cells do not considerably indent gels. Here, we evaluate the mechanical invasiveness of established breast and pancreatic cell lines and of tumor-cells from fresh, suspected pancreatic cancer tumors. The mechanical invasiveness matches the in vitro metastatic potential in cell lines as determined with Boyden chamber assays. Moreover, the mechanical invasiveness directly agrees with the clinical histopathology in primary-site, pancreatic-tumors. Thus, the rapid, patient-specific, early prediction of metastatic likelihood, on the time-scale of initial resection/biopsy, can directly affect disease management and treatment protocols.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is currently the second cause of death worldwide. Cancer-related deaths are predominately due to uncontrolled tumor progression, resulting in local or metastatic spread.34 The strongest prognostic predictors of metastatic risk in various cancer types are tumor size, lymph node status, histological grade, number of identifiable mitoses, by immunohistochemistry (e.g. Ki 67 staining) and the presence of necrosis in the specimen; those predictors are, however, not infallible. For example, approximately 30% of patients with breast cancer will develop metastases with apparently negative axillary lymph nodes.43 Rapidly progressing tumors require prompt and aggressive action, yet treatment protocols are determined by likelihood for metastases. Recurrence risk is currently predicted by lymph-node status, tumor size, histopathology and genetic testing, however, these are not infallible and results may require days/weeks. As the standard grading system and pathological tools are limited in their ability to rapidly and accurately predict the biological behavior of tumor-cells, the likelihood for metastatic progression or local recurrence are largely indeterminable, especially at the time of surgery.
The clinical gold-standard for tumor staging is histopathological examination, which is then used to tailor treatments. Pathology results in a busy cancer medical center may take several weeks; histological grading of tumor samples entails tissue fixation and manual imaging and analysis. Automated histopathological analysis of fixed tissue slices may accelerate results,16 yet accuracy is challenging and even novel deep-learning approaches have achieved only 67% agreement with the results of standard, manual histopathology.42 This time frame may cause substantial distress for the patients, and theoretically may degrade their immune defense and the healing process. Patient-specific genetic biomarkers are potentially more accurate, but these requires pre-identified reliable indicators and disease-specific genetic markers that are currently unavailable,32,43 e.g. in pancreatic cancer. In practice, the sensitivity and specificity of individual markers can vary widely, being affected by various physiological and pathological factors. Thus, a patient specific, quantitative, and rapid approach is critically needed for accurate prediction of the likelihood for metastasis, or metastatic risk, preferentially based only on the primary tumor, this may guide the extent of surgery and the use surgical technologies.
A critical step in metastatic initiation is the invasion of tumor-detached cancer cells through varying dense, surrounding tissues and environments, which includes interaction with different types of cells and extracellular matrix. To traverse their surroundings, the invading cancer cells must change morphology and apply forces.19,21,22 We and others have shown that invasive, metastatic cells are dynamically softer both internally14 and externally8,15,37 as compared to non-invasive and benign cells, yet can also restructure and apply stronger forces when advantageous.20,21,26,28 The intracellular and extracellular mechanics play an essential role in the cell dynamics, associated with invasion.13
Several in vitro assays have been designed to determine cell invasiveness and migratory capacity based on the cell deformability, yet variability in cell responses (related to tumor and cell line heterogeneity), assay complexity and extended assay times have prohibited translation into in vivo systems and the clinic. The in vitro metastatic potential (MP) of cells is commonly evaluated using transwell assays (Boyden chambers), however, those assays typically require serum starvation and an overall of 72–96 h; during the prolonged times, longer than cell doubling-times, fresh tumor-cells can undergo changes that may affect assay results. Boyden chamber migration/invasion experiments have been used with Matrigel coating2 to make them more physiologically relevant, yet as Matrigel batches are highly variable,33 ability to universally correlate the results to MP is limited. Similar issues with long-term changes in cells during extended experiment times occur in recently developed three-dimensional (3D), multicellular, tumor spheroid invasion assays,40,41 and while those mimic aspects of the in vivo tumor structure and function, generated spheroids are not robustly uniform.36 Cell culture in 3D promotes many biological relevant functions not observed in 2D,23 and gel invasion assays have been developed that utilize physiologically relevant collagen29 or gelatin5 matrices; the clinical relevance of those assays has yet to be established. More recently, 3D microfluidic-based approaches have been developed to allow control of the architecture and composition of the cancer-cell microenvironment.11 Those microfluidic assays have demonstrated higher cell-invasion rates than in Boyden chamber39 and are useful for single-cell invasion studies.17 Using a 3D microfluidic system, the invasiveness of breast cancer cell lines was quantitatively determined with results comparable to those of Boyden chambers assays, yet obtained more rapidly (after 24 h); preparatory cell starvation is required in both assays. The speed of result availability in those systems is currently tens of hours, and even then only few cells are evaluated in some cases45; tumor heterogeneity requires evaluation of a wide cell population to identify important outliers.
Here, we propose a clinically translatable approach based on evaluation of the in vitro mechanobiology of cells to identify the in vivo metastatic risk of a sample,44 and demonstrate direct agreement with the clinical histopathology; the system is not intended to model the in vivo environment, but to evaluate sampled cell invasive capacities. We have recently shown that metastatic cancer cells from breast cancer cell-lines will indent synthetic impenetrable gels within 1 h from seeding while benign cells do not significantly indent10,21,28 and the indenting breast-cancer cells are also the cell subpopulation that can traverse a Boyden chamber membrane.4 The current work shows that our ‘mechanical invasiveness’ assay may be applicative across different cancer types (as demonstrated on breast and pancreatic cancer cell lines), and directly agrees with clinical histopathology in fresh samples from primary pancreatic tumors; thus can be used to provide a rapid, early prognosis of metastatic risk.
Materials and Methods
Cell Lines
We have used ten commercially-available human epithelial cell lines from breast and pancreatic cancers, collected from primary site or from metastatic sites (all cells from American Tissue Culture Collection, ATCC, Manassas, VA). The breast cell lines were: cancerous with high MP (MDA-MB-231) and low MP (MDA-MB-468 and MCF7) from pleural effusion, and benign cells (MCF-10A) as control. The pancreatic cell lines were: Mia-Paca2, Panc1 and BxPC-3 (from primary site), AsPc1 (from ascites metastasis), Capan1 (from liver metastasis), SW1990 (from spleen metastasis).
Cell Culture
Cell lines were cultured in their appropriate media, as recommended by ATCC. For the MDA-MB-231, MDA-MB-468, MCF7, Mia-Paca2 cell lines we used Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen life technologies, Carlsbad, CA) supplemented with 10 vol% fetal bovine serum (FBS) (ThermoFisher Scientific, Waltham, MA), and 1 vol% each of l-glutamine, penicillin-streptomycin, and sodium pyruvate (all from Biological Industries, Kibbutz Beit Haemek, Israel). For the AsPC-1, Capan-1, BxPC-3, Panc1, SW1990 cell lines we used RPMI-1640 Medium (Biological Industries, Kibbutz Beit Haemek, Israel) supplemented with 10%vol. FBS (ThermoFisher Scientific, Waltham, MA), 1%vol. of penicillin-streptomycin (Biological Industries, Kibbutz Beit Haemek, Israel), 0.46%vol. d-Glucose solution, 1%vol. HEPES solution, and 0.66%vol. sodium bicarbonate solution (all from Sigma, St Louis, MO). Cells were cultured at 37 °C, 5% CO2, and high humidity. No mycoplasma testing was done. Cells were frozen at low passages from ATCC stock (i.e. 3–5), and for experiments cells were thawed and used in passages 7–30 from the ATCC stock.
Fresh Tumor Cells
To validate clinical relevance we have tested pancreatic tumor-samples from human subjects. Suspected tumors were resected according with all relevant ethical regulations, after obtaining informed consent from volunteers at the Rambam Health Care Campus at Haifa, Israel (Institutional Review Board, IRB approval number: RMB-0285-14). Non-edge, tissue samples not required for histopathological examination were transported and stored at 4 °C in histidine-tryptophan-ketoglutarate (HTK) preservation solution (Biological Industries, Israel). The samples were evaluated within 12 h from resection. Each sample was measured in three dimensions by caliper, weighed, photographically documented and given a running number for archiving purposes. Cells were isolated from minced tissue samples using the Tumor dissociation kit (Miltenyi Biotec, Auburn, CA) as demonstrated in Fig. 1. In short, enzymatic degradation was performed at 37 °C with shaking, during 2 h. The cell-extract was passed through a 100 µm cell-strainer (Corning Inc., Corning, NY) to separate non-degraded tissue pieces. The cells were then concentrated by centrifugation and non-nucleated cells (e.g. red blood cells) were removed by 4 min incubation with cell lysis buffer (Roche Diagnostics, Germany). Finally, the suspected cancer cells were transferred to the RPMI-1640 culture media (Biological Industries, Israel) contains 10% FBS (Hyclone, MA), 1% penicillin-streptomycin (Biological Industries, Israel) and immediately seeded on the gel for evaluation; cells were discarded after use.
Cell extraction from tumors. Fresh tissue samples are transported to the lab in HTK solution. Cells are extracted from freshly resected tissue sample by enzymatic degradation. Degradation is performed at 37 °C under shaking and any non-degraded tissue fragments are removed by 100 µm cell-strainer. Blood cells are then removed by lysis buffer. The process of cell extraction requires 2.5–3 h. Collected cells are immersed in cell-growth media and immediately seeded on polyacrylamide gels for indentation measurements.
Polyacrylamide Gel Preparation
The polyacrylamide (PAM) gels were prepared at physiological stiffness (Young’s modulus 2.4 kPa) according to our established protocol.4,10,21,26,27 In short, gels were prepared on glass number 5 (thickness 0.5-0.6 mm) coverslips, 30 mm diameter (Menzel, Germany), inside a plastic frame (Gene Frame, 25 μL, 10 × 10 mm; ABgene Thermo Scientific, Waltham, MA) using a constant ratio of 179:1 of the monomers Acryl:cross-linker BIS-acrylamide (both from Bio-rad, Israel) in distilled water; specifically, we used34µl of 40 vol.% acrylamide and 3.8 µl of 2 vol.% BIS acrylamide in 203 µl of distilled water; Polymerization was initiated with 1:200 vol. ammonium persulfate and catalyzed with 1:500 vol. tertiary aliphatic amine N,N,N’,N’-tetramethylethylenediamine, TEMED (both from Sigma, St Louis, MO). The above recipe produced a gel with Young’s modulus of 2.4 ± 0.14 kPa, as was previously determined at our laboratory,21,28 and re-confirmed here. Red (excitation/emission 580/605 nm) fluorescent carboxylated polystyrene particles (Molecular probes, Invitrogen life technologies, Carlsbad, CA), 200 nm in diameter, were added to the gel and brought to just below its surface by performing slow gelation at 2 °C under 300 g centrifugation for 30 min. Finally, the surface of the gel was activated with Sulfo-SANPAH (Pierce, Thermo Scientific, Waltham, MA), washed with HEPES, and coated with 5 µg/ml rat tail collagen type I (Sigma, St Louis, MO) for cell adhesion.
Gel mechanics were evaluated with a TA Instruments AR-G2 magnetic bearing, shear rheometer (New Castle, DE) with a 2-cm parallel stainless-steel plate fixture. Gelation is induced on the rheometer plate to avoid slip, and the average shear, storage and loss moduli, respectively, G*, G′ and G″, were measured in a time-sweep under 0.5% strain and angular frequency of 3.14 rad/s, to track and verify polymerization.1 The Young’s modulus was the determined by: E = 2G′(1 + ν), as the G* ≈ G′ in the elastic gel, and the Poisson’s ratio, ν, is 0.49 for PAM gels. The gels were produced with a Young’s modulus of 2.4 ± 0.14 kPa and stiffness exhibited small variation and high reproducibility between batches.
Cell Viability and Nuclei Staining
Calcein-AM fluorometric assay (BioVision, USA) was used for viability staining. Calcein AM is a non-fluorescent, hydrophobic compound that easily penetrates intact and live cells producing a fluorescently labeled cell cytoplasm (excitation/emission 485/530 nm) 30 min after staining. Hoechst assay (Sigma, St. Louis, MO) was used to stain nuclei by incubating for 0.5–1 h and imaging fluorescently labeled nuclei (excitation/emission 346/460 nm).
Microscopy and Imaging
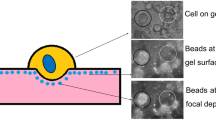
We seeded 300,000 cells on each gel within the appropriate cell media, resulting in an average of 30 ± 10 viable cells per field-of-view (area of 0.016 mm2). Cells were seeded so they were closely adjacent yet in a monolayer without overlap. We have previously shown that single, well-spaced or closely adjacent cells demonstrate similar overall percentage of indenting cells, yet different distributions of indentation depths.28 The closely adjacent cells provided a distinction between low and high metastatic potential cell samples, and also provided higher statistics per sample, and are thus used here. The imaging was done with an inverted, epifluorescence Olympus IX81 microscope, using a 60×/0.7NA differential interference contrast (DIC, Nomarsky optics) air-immersion, long working-distance objective lens. The cells were maintained in 37 °C, 5% CO2, and high humidity (90%) throughout the entire experiment to sustain their viability. Imaging and indentation depth measurements were performed from 45 min to 2 h after seeding, as per our previous studies,21,28 which allows collection of sufficient statistic and is a clinically relevant time-scale.
In each gel we imaged 9–10, randomly chosen, fields-of-view, and each experiment included 2–3 separate gels. At each randomly chosen location on the gel, at least 3 images were taken (Fig. 2): (i) a DIC image of the cell morphology on the gel, (ii) a fluorescence image of the particles embedded at the focal plane of the gel surface, and (iii) a series of fluorescence images at the focal depth to which particles had been displaced, to identify each indenting cells’ depth; the focal height of each image was automatically recorded upon image capture using the automated microscope and lens-controller. The series included images at 5–6 focal depths below the gel height, where 1–8 indenting cells were in focus at each depth. We have previously shown that the indentation depth is independent of the cell size and other cell-scale metrics,10,21 hence those are not required for the assay and are not determined.
Mechanical invasiveness assay. Experimental procedure includes cell-seeding on polyacrylamide (PAM) gel with fluorescent nano-particles embedded in its surface. Following seeding and cell attachment (< 1 h), a subpopulation of the cells indents the gels. Images on sides are: (top) DIC images of cell morphology; (middle) fluorescent images of particles at gel surface (0 µm); (bottom) fluorescent images of particles moved to 8.2 µm depth below the gel surface by indenting cells. Images on bottom are full-frames of the field of view from which the magnified images were obtained. The indentation depth (∆L) is the focal depth difference between the gel surface and the lowest in-focus particle-plane.
Indentation Depth Determination
The focal height of each acquired image is automatically recorded by a custom microscope-control module in MATLAB 2017b (The Mathworks, Nattick, MA) and exported for each field-of-view. Following the experiment, images were analyzed using a second custom-designed module in MATLAB to determine the number of viable and indenting cells, as well as the indentation depth of each cell; the percent of indenting cells is out of the total adhered and viable cells. Specifically, in each field-of-view, the cells that have attained a specific indentation depth (are in focus in the image) marked by the user in the MATLAB module. Then the indentation depth of each marked cell is automatically calculated and recorded to a file and a count of the indenting cells is generated. The indentation depth was calculated by the difference in focal height of the gel surface (undisturbed gel) and the lowest focal plane where particles are identified to be in focus, i.e. below the specific indenting cell. In addition, the analysis module automatically counts the overall number of cells in a field-of-view and identifies the viable cells and their percentage, as previously published.3
Boyden Chamber, Trans-Well Migration Assay
We have determined the percentage of cancer cells from all cell lines that cross through 8 µm Boyden chamber membranes. In short, cells were serum starved overnight and 10,000 cells were seeded in 100 µl of serum-free cell-type specific media in the upper compartment of transwell inserts (EDM Milli-pore, Billerica, MA) with 8 µm pores coated with 5 µg/ml rat tail collagen type I (Sigma, St Louis, MO), overnight at 4 °C. Serum-full growth media was placed in the lower chamber, serving as a chemoattractant. Cells were incubated in the wells for 72 h at 37 °C and 5% CO2. The cells from the upper and lower compartments were collected, by incubation with 200 and 300 µl respectively of trypsin-EDTA solution A (0.25%; Biological Industries, Cromwell, CT) for 5 min at 37 °C and under 5% CO2. The trypsin was neutralized by adding 0.8 and 1.2 ml of growth media to the upper and lower compartments, respectively, and the cells were counted using hemocytometer. For each cell type 6–9 independent experiments were performed, and percentage of trespassing cells determined.
Statistical Analysis
Significance of variations between cell-lines were determined using the general, linear mixed model, a multivariate regression method for the analysis of variance (ANOVA), with a p value < 0.05 (see Table S1). In addition, the Spearman rank correlation coefficient, was used to determine the strength and direction (+/-) of association between the results of the mechanical invasiveness and the Boyden chamber assays. Calculations were performed using Excel (MicroSoft, Redmond, WA) or MedCalc2018 (MedCalc Software, Ostend, Belgium). Automated k-means clustering using Lloyd’s algorithm25 was performed in MATLAB to demonstrate separation of results to non-invasive and invasive subpopulations. Following indication of number of clusters (i.e. 2), the algorithm automatically determines the centroid of each cluster and the included samples.
Results
We determine the mechanical invasiveness of a cell sample by evaluating the amounts of indenting cells and their attained depths (Fig. 2). Cells are seeded on physiological-stiffness synthetic polyacrylamide gels that are non-degradable and with sub-micron pores; i.e. gels are impenetrable even to highly invasive cells and chosen 2.4 kPa stiffness is on the scale of soft tissue. Within 1 h of seeding, cell attach and we observe a subset of the adhered cells that forcefully indent the 2.4 kPa gel to depths on the order of cell size (Fig. 2); overall cell viability on gels, including indenting and non-indenting cells, is 96-99% in all the evaluated cell lines (Fig. S1). Imaging random regions on the gel surface, using a fluorescent microscope, allows us to quantify the amounts of gel-indenting (viable) cells and the attained indentation depth of each cell; we use a semi-automated custom module to evaluate these measures. The morphology of indenting/non-indenting cells is indistinguishable by light microscopy, being predominately rounded as expected on soft substrates21,28 and also characteristically observed during tumor cell invasion through three-dimensional collagen-based matrices.12
Using the above assay, we have evaluated the mechanical invasiveness of established pancreatic (n = 6) and breast (n = 4) cell lines. We observe increasing amounts of indenting cells (Fig. 3a) and their average, attained indentation depth (Fig. 3b), which agrees with their ATCC and literature established MP (Table 1) and with the in vitro MP as evaluated via Boyden chamber assay (Fig. 3c); the cell lines, all adherent epithelial cells, were collected from primary or metastatic sites. The indentation depths attained by individual, yet closely situated cells exhibit a Gaussian-like distribution (Fig. 4). We observe a significant difference (p < 0.0001) between the means of three groups: benign/non-metastatic, low MP and high MP cells.
(a–b) Invasiveness of breast (green) and pancreatic cancer (gray) cell-lines on 2.4 kPa stiffness polyacrylamide-gel. Increased metastatic potential (MP) leads to B, larger fraction of indenting cells; (c) larger, average indentation depths (n≥9). (c) Cells trespassing 8-µm pore Boyden-chambers (n≥6). (d) Mechanical invasiveness (indentation capacity) and Boyden-chamber assay agree in percentage of indenting cells and in (e) indentation depths. Dashed lines are guides to the eye. Statistical significance calculated using one-way ANOVA with Tukey test is provided in Table S1; data represented as mean ± standard error of the mean (s.e.m.).
All evaluated breast and pancreatic cancer cell lines display Gaussian-like distribution of indentation depths on 2.4 kPa PAM gels. Dotted vertical lines indicate average indentation depths for (a) non-metastatic cells (2.67 ± 0.76 µm), (b) low to medium metastatic potential (MP) cells (4.37 ± 0.89 µm) and (c) high MP cells (5.3 ± 1.09 µm). The average values (± s.e.m) of indentation depths for 3 groups of cells are significantly different (p < 0.001).
We have validated the in vitro MP of the cell lines with the commonly used Boyden chamber, transwell migration and invasion assay; the method is simple yet takes 48–96 h and requires serum starvation of cells. We have recently shown a direct link between the sub-population of invasive cells that trespass the Boyden chamber membrane and those that indent the gels.4 As expected, we observe an increasing amounts of cells that trespass the Boyden chamber membrane (8 µm pores), within 72 h from seeding (Fig. 3c), with increasing, established MP (Table 1). We note, however, that not all cell-lines can be differentiated by this method. For example, differences between the following groups were not statistically significant: benign cells do not significantly differ from low MP cancer cells, low MP cells from the same cancer type (i.e. breast or pancreas) do not differ, and high MP pancreatic cancer cells are not statistically different (Table S1). We observe that in those cases the mechanical invasiveness was statistically different and distinguished between the cell lines with no, low, medium, or high MP; such categorization of MP is critical for clinical and research applications.
The results of the 72-h Boyden-chambers assay correlate with both measures of the 2-h gel-indentation, mechanical invasiveness assay, i.e. the amounts of indenting cells (Fig. 3d) and their attained depths (Fig. 3e); both measures are potential indicators of the MP. Specifically, the percentage of transwell-trespassing cells demonstrate Spearman-Rank correlation coefficients of 0.73 (p < 0.02) and 0.92 (p < 0.0001), respectively, with the percentage of indenting cells (Fig. 3d) and with the indentation depths (Fig. 3e). We note that the correlation between the Boyden and mechanical invasiveness assays is lower in the percentage of indenting cells due to differences between the two cancer types (see x-axis shift in Fig. 3d). The differences result from a larger relative number of breast cancer cells that trespass the Boyden chamber membranes, as highlighted and corroborated by the mechanical invasiveness measure. Thus, the mechanical invasiveness of pancreatic and breast cancer cell lines matches literature-established and Boyden-chamber in vitro MP (Fig. 3c), while being rapid (hours vs. days) and also demonstrating unique cancer-type-dependent differences (Fig. 3d–e). We hereafter define higher mechanical invasiveness by combined increase in amounts of indenting cells and the average attained indentation depths (Fig. 5), i.e. cells with high MP exhibit more indenting cells and/or reach deeper depths.
Mechanical invasiveness of cell-lines and pancreatic tumor samples on 2.4 kPa PAM gels. (a) Amounts of indenting cells and depths, together the mechanical invasiveness, in breast (filled triangle) and pancreatic (filled diamond) cell-lines agree with metastatic potential; shade from light to dark indicates literature-established increase in metastatic potential. (b) Suspected fresh samples from pancreas (filled circle). Invasive adenocarcinoma demonstrated higher mechanical invasiveness than benign/precancerous (IPMN) and normal samples; increase in invasiveness indicated by change in shade. Asterisk indicates four samples from tumor-adjacent sites in different volunteers that are at the origin, zero indentation. (c) K-means cluster analysis of pancreatic tumor-samples (filled circle) together with breast (filled triangle) and pancreatic (filled diamond) cell-lines demonstrates separation between metastatic and non-metastatic cells. Data represented as mean ± s.e.m.
To demonstrate the clinical relevance and applicability of our approach, we have evaluated the mechanical invasiveness of cells extracted from primary-site pancreatic tumor-samples from human subjects, using the clinical histopathology and patient outcome as the gold standard for comparison. We have collected samples from patients that did not exhibit metastasis upon initial presumed-cancer diagnosis and resection; this allows testing of early prediction of metastatic risk. We have extracted cells from primary-site tissue-samples (0.25–2 g) within hours from resection (Fig. 1); reliable results can be obtained with as little as 30,000 cells in the current setup. In benign or pre-malignant samples (as determined later by pathological examination), only a low percentage of cells (< 15%) indent the gels and those attain small depths of < 4 µm (Fig. 5b); results in fresh tumor samples match benign cell lines (Fig. 5a). Moreover, in normal samples from non-tumor sites in the pancreas we observe little mechanical invasiveness (near no-indentation, Fig. 5b).
Tumor sample results agree with clinical diagnosis/prognosis and match cell lines. In cells from tumor samples we observe significant indentions. For example, the sample primay-tumor01 (Table 2) exhibited 39.7 ± 2.2% indenting cells that reached average depths of 4.8 ± 0.4 µm with a long-tail distribution (Fig. 5b and inset); results were on scale of high MP pancreatic cancer cell lines. Consequently, the donor of primay-tumor01 was clinically diagnosed with aggressive pancreatic cancer: moderately differentiated, Stage 3 adenocarcinoma, and liver metastasis developed within 1.5 months of surgery; the donor unfortunately passed away 4.5 months later. In contrast, the sample primary-tumor02 exhibited 14 ± 1.3% indenting cells with average depths of 2.2 ± 0.2 µm with Gaussian-like distribution (Fig. 5b and inset), similar to benign/non-invasive cell lines (Fig. 5a). The donor was unsurprisingly diagnosed as having a pre-cancerous lesion. Our results, obtained within ~2 h from the primary-site tumor sample, thus match the histopathology and agree with the clinical diagnosis/prognosis in predicting the patient outcomes, e.g. rapid metastatic disease progression (Table 2). To generalize the prognostic capacity of the assay, we have applied an automated, machine learning, clustering analysis. We have performed k-means clustering on the mechanical invasiveness data set of the evaluated cell-lines and tumor-samples, defining two centroids (k = 2) and allowing automated segmentation. Figure 5c shows two well-separated groups (with 95% confidence levels) indicating the ability of the assay to distinguish, with the existing samples, benign and non-invasive samples from invasive and potentially metastatic samples.
Discussion
Using the mechanical invasiveness assay, i.e. measuring the percentage of indenting cells and their attained indentation depths, we were able rapidly and quantitatively distinguish between cancer cells that are benign/non-invasive or are likely to metastasize. As we have previously shown, widefield fluorescence microscopy provides the same mechanical invasiveness measures as confocal microscopy,10,21,28 yet is simpler to apply and readily available in most laboratory and clinical settings. We have validated our mechanical invasiveness results in vitro in breast cancer4,10,21,28 and pancreatic cancer cell lines with the commonly used Boyden chambers assay. An increase in mechanical invasiveness (amounts of indenting cells and their depths) matched the percentage of migrating cells; that is the indentation capacity agrees with the migratory capacity of the cells of the same cancer type. Specifically, results matched in benign, non-invasive, low MP and high MP cells, yet differences between cell types were more statistically significant in the mechanical invasiveness assay. We note that the attained indentation depths, on the 2.4 kPa gel, vary similarly with pre-determined MP for cell lines from both evaluated cancer types (Fig. 3e). In contrast, combined analysis of the amounts of indenting and migrating cells (Fig. 3d) reveals a separation between breast and pancreatic cancer cell lines; i.e. higher relative invasiveness is expected in the breast cells. Aptly, highly metastatic breast and pancreatic cell lines injected into mice induced different rates of in vivo metastasis occurrence, respectively being 97 and 83%.38
The mechanical invasiveness assay provides results of cell invasive capacity within 1–2-h from seeding, demonstrating that rapid mechanobiological interactions of cells can be utilized to stratify the metastatic potential of cells within a (tissue) sample. The rapidity of obtained results, use of standard cell-culture media (no need for serum starvation),4 and reliance on widefield microscopy may revolutionize clinical diagnostics in oncology and the ensuing disease management. We note that the Boyden chamber results (obtained in 48–96 h) and the 2-h mechanical invasiveness assay, potentially evaluate different stages of the invasive process than, yet there is agreement in the relative invasiveness of established cell lines. Thus, the mechanical invasiveness assay rapidly probes the invasive capacity of the sampled cells, which is utilized on different timescales in vivo, e.g. short timescales intra- and extra-vasation and longer time-scales invasion through tissue. The mechanical invasiveness assay concurrently evaluates large portions of the overall population of extracted cells, thus considering the wide heterogeneity of the tumor sample. This has been highlighted as a problem in single-cell evaluation methods in cancer research, which can affect results based on subsamples of the tumor.24
Pancreatic cancer is the fourth leading cause of cancer related mortality and predicted to become the second by 2030.31 Survival of pancreatic adenocarcinoma remains poor, although significant research focus is aimed at providing better patient care and improved overall survival. Recent neoadjuvant and adjuvant chemotherapy regimens improved median survival in resectable and borderline resectable patients to 15–32 months,35 with the 1-year survival peaking at 79.5% and then steeply declining to 50.1% in the second year; 5-year survival of pancreatic cancer remain dismal at 10%.6 The lethality of pancreatic cancer is attributed to the biological behavior of the disease, the lack of screening programs, and biological markers. The poor understanding of this complex and rapidly progressing disease has led to advanced pathological staging at first diagnosis and the reported grim prognosis. Interestingly, in 30–40% of newly diagnosed (stage 1–2) pancreatic cancer cases,7,30 the disease has yet to induce metastases or those are too small to be identified with current approaches. Existence of metastases reduces survival rates, and 90% of cancer related deaths are due to either metastatic or locally advanced disease.34 Choice of treatment is directly affected by the (predicted or estimated) biological behavior of the disease. Specifically, patients whose cancers would metastasize should go through aggressive chemotherapy while those whose cancers would remain localized should to go through an aggressive surgical approach such as extended and radical pancreatic resections and radiotherapy. Thus, it is critical to provide rapid, early prediction of metastatic likelihood already at first diagnosis of the primary tumor. The automated clustering analysis applied to the indentation results of the several cell lines and tumor samples evaluated here reveals two well-separated groups; results were verified, respectively, by the Boyden chamber and clinical histopathology and follow-up. The groups separate benign and non-invasive samples from invasive and potentially metastatic samples, with >95% confidence. our mechanical invasiveness assay could potentially provide a rapid, early prognosis platform (not requiring knowledge of tumor genetics or specific molecular markers or mimicking the natural physiological tumor environment) that can reinforce histopathology results and facilitate choice of patient-optimal treatment protocols.
To summarize, we have demonstrated the successful application of the mechanical indentation assay on pancreatic and breast cancer samples, yet as the mechanical invasiveness provides a non-genotype-based approach to evaluate metastatic capacity, the approach is expected to be cross-cancer relevant in various types of (solid) tumors. The (mechanical) invasiveness is likely an inherent capacity of the cells, being a composite of their dynamic ability to change shape, apply forces, and migrate; further samples from other cancers would be required for verification of generality. As the assay results are obtained on the time-scale of a typical medical procedure (i.e. 2–4 h), and may be applied with widefield (fluorescence) microscopy that is typically available in the majority of research lab and clinics/diagnostic centers, it is highly relevant for clinical practice. Clinical translation would require robust determination of the diagnostic/prognostic ranges in the mechanical invasiveness plot for each cancer type of interest. This would eventually facilitate standardization of the technique for simple and consistent use. Using the current protocols, results obtained by several different users (> 10) with varying backgrounds have been robustly consistent over a period of nearly a decade; i.e. no user bias is expected. Further automation of the assay and of the analysis, such as the clustering analysis using results from several users and centers will also facilitate wide clinical use. Importantly, the assay is designed to allow performance of all the accepted clinical tests, such as histopathological testing, e.g. as it does not require samples from the tumor edges. Early determination of metastatic likelihood directly from a sample of the primary tumor will have a significant impact on disease management and chosen treatment protocols. The mechanical invasiveness assay will initially likely be used as an adjuvant to the current, standard diagnostic/prognostic tests. This will allow to combine results from the clinical-standard heuristic approaches and long-term patient outcomes with the mechanical invasiveness assay results, in a Bayesian sense e.g. by developing machine learning modules to improve the accuracy of the assay and to automate the provided prognosis. Providing a rapid, accurate and quantitative measure for early prognosis of metastatic likelihood will allow physicians to apply patient-specific treatment protocols already during initial resection and will improve long-term outcome and treatment of cancer patients.
References
Abuhattum, S., and D. Weihs. Asymmetry in traction forces produced by migrating preadipocytes is bounded to 33. Med. Eng. Phys. 38:834–838, 2016.
Albini, A., and R. Benelli. The chemoinvasion assay: a method to assess tumor and endothelial cell invasion and its modulation. Nat. Protoc. 2:504–511, 2007.
Alvarez-Elizondo, M. B., C. W. Li, A. Marom, Y.-T. Tung, G. Drillich, Y. Horesh, S. C. Lin, G.-J. Wang, and D. Weihs. Micropatterned topographies reveal measurable differences between cancer and benign cells. Med. Eng. Phys. 75:5–12, 2020.
Alvarez-Elizondo, M. B., and D. Weihs. Cell-gel mechanical interactions as an approach to rapidly and quantitatively reveal invasive subpopulations of metastatic cancer cells. Tissue Eng. Part C Methods 23:180–187, 2017.
Artym, V. V., K. M. Yamada, and S. C. Mueller. ECM degradation assays for analyzing local cell invasion. In: Methods in Molecular Biology (Clifton, N.J.). 2009, pp. 211–219.
Carrato, A., A. Falcone, M. Ducreux, J. W. Valle, A. Parnaby, K. Djazouli, K. Alnwick-Allu, A. Hutchings, C. Palaska, and I. Parthenaki. A systematic review of the burden of pancreatic cancer in Europe: real-world impact on survival, quality of life and costs. J. Gastrointest. Cancer 46:201–211, 2015.
Casciani, F., G. Marchegiani, G. Malleo, C. Bassi, and R. Salvia. Pancreatic cancer in the era of neoadjuvant therapy: a narrative overview. Chirurgia (Bucur) 113:307, 2018.
Cross, S. E., Y. S. Jin, J. Rao, and J. K. Gimzewski. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2:780–783, 2007.
Deer, E. L., J. Gonzalez-Hernandez, J. D. Coursen, J. E. Shea, J. Ngatia, C. L. Scaife, M. A. Firpo, and S. J. Mulvihill. Phenotype and genotype of pancreatic cancer cell lines. Pancreas 39:425–435, 2010.
Dvir, L., R. Nissim, M. B. Alvarez-Elizondo, and D. Weihs. Quantitative measures to reveal coordinated cytoskeleton-nucleus reorganization during in vitro invasion of cancer cells. New J. Phys. 17:043010, 2015.
Eslami Amirabadi, H., S. Saheb Ali, J. P. Frimat, R. Luttge, and J. M. J. Den Toonder. A novel method to understand tumor cell invasion: integrating extracellular matrix mimicking layers in microfluidic chips by “selective curing”. Biomed. Microdevices 19:1–11, 2017.
Friedl, P., and K. Wolf. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer 3:362–374, 2003.
Friedl, P., and K. Wolf. Plasticity of cell migration: a multiscale tuning model. J. Cell Biol. 188:11–19, 2010.
Gal, N., D. Lechtman-Goldstein, and D. Weihs. Particle tracking in living cells: a review of the mean square displacement method and beyond. Rheol. Acta 52:425–443, 2013.
Guck, J., S. Schinkinger, B. Lincoln, F. Wottawah, S. Ebert, M. Romeyke, D. Lenz, H. M. Erickson, R. Ananthakrishnan, D. Mitchell, J. Kas, S. Ulvick, and C. Bilby. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 88:3689–3698, 2005.
Gurcan, M. N., L. E. Boucheron, A. Can, A. Madabhushi, N. M. Rajpoot, and B. Yener. Histopathological image analysis: a review. IEEE Rev. Biomed. Eng. 2:147–171, 2009.
Holle, A. W., N. Govindan Kutty Devi, K. Clar, A. Fan, T. Saif, R. Kemkemer, and J. P. Spatz. Cancer cells invade confined microchannels via a self-directed mesenchymal-to-amoeboid transition. Nano Lett. 19:2280–2290, 2019.
Holliday, D. L., and V. Speirs. Choosing the right cell line for breast cancer research. Breast Cancer Res. 13:215, 2011.
Koch, T. M., S. Munster, N. Bonakdar, J. P. Butler, and B. Fabry. 3D Traction forces in cancer cell invasion. PLoS ONE 7:e33476, 2012.
Kraning-Rush, C. M., J. P. Califano, and C. A. Reinhart-King. Cellular traction stresses increase with increasing metastatic potential. PLoS ONE 7:e32572, 2012.
Kristal-Muscal, R., L. Dvir, and D. Weihs. Metastatic cancer cells tenaciously indent impenetrable, soft substrates. New J. Phys. 15:035022, 2013.
Levental, K. R., H. Yu, L. Kass, J. N. Lakins, M. Egeblad, J. T. Erler, S. F. T. Fong, K. Csiszar, A. Giaccia, W. Weninger, M. Yamauchi, D. L. Gasser, and V. M. Weaver. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139:891–906, 2009.
Li, X., A. V. Valadez, P. Zuo, and Z. Nie. Microfluidic 3D cell culture: potential application for tissue-based bioassays. Bioassays. 4(12):1509–1525, 2012.
Liang, S. B., and L. W. Fu. Application of single-cell technology in cancer research. Biotechnol. Adv. 35(4):443–449, 2017.
Lloyd, S. Least squares quantization in PCM. IEEE Trans. Inf. Theory 28:129–137, 1982.
Massalha, S., and D. Weihs. Metastatic breast cancer cells adhere strongly on varying stiffness substrates, initially without adjusting their morphology. Biomech. Model. Mechanobiol. 16:961–970, 2017.
Merkher, Y., M. B. Alvarez-Elizondo, and D. Weihs. Taxol reduces synergistic, mechanobiological invasiveness of metastatic cells. Converg. Sci. Phys. Oncol. 3:044002, 2017.
Merkher, Y., and D. Weihs. Proximity of metastatic cells enhances their mechanobiological invasiveness. Ann. Biomed. Eng. 45:1399–1406, 2017.
Nyström, M., G. Thomas, M. Stone, I. Mackenzie, I. Hart, and J. Marshall. Development of a quantitative method to analyse tumour cell invasion in organotypic culture. J. Pathol. 205:468–475, 2005.
Peixoto, R. D., C. Speers, C. E. McGahan, D. J. Renouf, D. F. Schaeffer, and H. F. Kennecke. Prognostic factors and sites of metastasis in unresectable locally advanced pancreatic cancer. Cancer Med. 4:1171–1177, 2015.
Rahib, L., B. D. Smith, R. Aizenberg, A. B. Rosenzweig, J. M. Fleshman, and L. M. Matrisian. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 74:2913–2921, 2014.
Riihimaki, M., H. Thomsen, A. Hemminki, K. Sundquist, K. Hemminki, M. Riihimäki, H. Thomsen, A. Hemminki, K. Sundquist, K. Hemminki, M. Riihimaki, H. Thomsen, A. Hemminki, K. Sundquist, and K. Hemminki. Comparison of survival of patients with metastases from known versus unknown primaries: survival in metastatic cancer. BMC Cancer 13:36, 2013.
Sieuwerts, A. M., J. G. Klijn, and J. A. Foekens. Assessment of the invasive potential of human gynecological tumor cell lines with the in vitro Boyden chamber assay: influences of the ability of cells to migrate through the filter membrane. Clin. Exp. Metastasis 15:53–62, 1997.
Sleeman, J. P., I. Nazarenko, and W. Thiele. Do all roads lead to Rome? Routes to metastasis development. Int. J. Cancer 128:2511–2526, 2011.
Suker, M., B. R. Beumer, E. Sadot, L. Marthey, J. E. Faris, E. A. Mellon, B. F. El-Rayes, A. Wang-Gillam, J. Lacy, P. J. Hosein, S. Y. Moorcraft, T. Conroy, F. Hohla, P. Allen, J. Taieb, T. S. Hong, R. Shridhar, I. Chau, C. H. van Eijck, and B. G. Koerkamp. FOLFIRINOX for locally advanced pancreatic cancer: a systematic review and patient-level meta-analysis. Lancet Oncol. 17:801–810, 2016.
Sung, K. E., and D. J. Beebe. Microfluidic 3D models of cancer. Adv. Drug Deliv. Rev. 79–80:68–78, 2014.
Swaminathan, V., K. Mythreye, E. T. O’Brien, A. Berchuck, G. C. Blobe, and R. Superfine. Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res. 71:5075–5080, 2011.
Teng, Y., X. Xie, S. Walker, D. T. White, J. S. Mumm, and J. K. Cowell. Evaluating human cancer cell metastasis in zebrafish. BMC Cancer 13:453, 2013.
Toh, Y. C., A. Raja, H. Yu, and D. Van Noort. A 3D microfluidic model to recapitulate cancer cell migration and invasion. Bioengineering 5:10, 2018.
Truong, H. H., J. de Sonneville, V. P. S. S. Ghotra, J. Xiong, L. Price, P. C. W. W. Hogendoorn, H. H. Spaink, B. van de Water, and E. H. J. J. Danen. Automated microinjection of cell-polymer suspensions in 3D ECM scaffolds for high-throughput quantitative cancer invasion screens. Biomaterials 33:181–188, 2012.
Vinci, M., C. Box, and S. A. Eccles. Three-dimensional (3D) tumor spheroid invasion assay. J. Vis. Exp 2015. https://doi.org/10.3791/52686.
Wei, J. W., L. J. Tafe, Y. A. Linnik, L. J. Vaickus, N. Tomita, and S. Hassanpour. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci. Rep. 9:3358, 2019.
Weigelt, B., J. L. Peterse, and L. J. van’t Veer. Breast cancer metastasis markers and models. Nat. Rev. Cancer 5:591–602, 2005.
Weihs, D., and Y. Merkher. A device and method for determining cell indention activity, Patent pending. Patent: PCT/IL2019/050463, 2019.
Yankaskas, C. L., K. N. Thompson, C. D. Paul, M. I. Vitolo, P. Mistriotis, A. Mahendra, V. K. Bajpai, D. J. Shea, K. M. Manto, A. C. Chai, N. Varadarajan, et al. A microfluidic assay for the quantification of the metastatic propensity of breast cancer specimens. Nat. Biomed. Eng. 3:452–465, 2019.
Acknowledgments
The work was partially funded by the Technion Internal Elias Fund for Medical Research and by Polak Fund for Applied Research, and by the Ber-Lehmsdorf Foundation and the Gerald O. Mann Charitable Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Konstantinos Konstantopoulos oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Merkher, Y., Horesh, Y., Abramov, Z. et al. Rapid Cancer Diagnosis and Early Prognosis of Metastatic Risk Based on Mechanical Invasiveness of Sampled Cells. Ann Biomed Eng 48, 2846–2858 (2020). https://doi.org/10.1007/s10439-020-02547-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-020-02547-4