Abstract
Development of insecticide resistance in insect populations is a major challenge to sustainable agriculture and food security worldwide. Buprofezin, one of the commonly used chitin synthesis inhibitors, has severely declined its control efficacy against the brown planthopper (BPH, Nilaparvata lugens), a devastating rice insect species. To date, however, mechanism of buprofezin resistance in target pests remains elusive. We conducted a long-term (25 years from 1996 to 2020) and large geographical scale (11 provinces and cities in China) resistance monitoring program for buprofezin in BPH, a notorious pest of rice crop in East and Southeast Asia. BPH rapidly developed resistance with > 1,000-fold resistance being detected in nearly all the field populations after 2015. Using the bulk segregant mapping method, we uncovered a novel mutation (G932C) in chs1 gene encoding chitin synthase 1 from a near isogeneic buprofezin-resistant (> 10,000-fold) strain harboring recessive, monogenic resistance. Using CRISPR/Cas9-based genome-modified Drosophila melanogaster possessing the same mutation as a model, we found that the G932C mutation was not only responsible for buprofezin resistance but also conferred a cross-resistance to cyromazine, an insect molting disruptor, on which the mode of action is largely unknown. Taken together, our study for the first time revealed the molecular mechanism conferring buprofezin resistance in BPH and implicated that cyromazine also targets chitin biosynthesis to confer its toxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Key messages
-
A 25-year monitoring program revealed widespread resistance to buprofezin in N. lugens in China
-
Buprofezin resistance was linked to a G932C mutation of chs1 gene encoding chitin synthase 1
-
Genome-modified Drosophila with the same mutation showed resistance to buprofezin and cyromazine
-
The G932C mutation likely played major roles conferring buprofezin resistance in N. lugens
-
Cyromazine, an insect molting disruptor, may also target chitin synthesis to confer its toxicity
Introduction
The brown planthopper (BPH, Nilaparvata lugens) is one of the most devastating insect pests of the rice crop in the temperate and tropical regions of East and Southeast Asia. The BPH not only causes direct damage through feeding but also transmits serious rice virus diseases. In Asia, it has been estimated that BPH alone causes an economic loss of more than $300 million annually (Wang et al. 2008b). In China, it has been estimated that extremely high levels of imidacloprid resistance in BPH caused a yield loss of 3.8 billion kilograms of rice in 2005 alone (Wang et al. 2008c, d). Thus, successful control of rice planthoppers is one of the key factors to ensure the yield and quality of rice, which represents the major staple crop for about 50% of the world’s population (Khush 1999).
The management of BPH primarily relies on the use of agrochemicals. Buprofezin is a thiadiazine compound (Group 16, IRAC grouping system) that acts as a chitin synthesis inhibitor (Insecticide Resistance Action Committee 2021). It is the only chitin synthesis inhibitor targeting hemipteran insect pests (Pener and Dhadialla 2012) and, therefore, is of great importance for controlling BPH. Indeed, buprofezin has been used to control BPH since the 1980s (Wang et al. 2008c) and remained its good efficacy for about 20 years as an alternative of many other insecticides including the neonicotinoid insecticides, such as imidacloprid. However, due to the outbreak of the BPH populations with extremely high resistance to imidacloprid in 2005, buprofezin has become the most extensively used insecticide to control BPH since then. As a result, BPH rapidly developed resistance to buprofezin and has shown extremely high levels of resistance in several geographical regions of China since 2013 (Wang et al. 2008a; Wu et al. 2018).
Early studies on mechanisms of buprofezin resistance in rice planthoppers focused on the small brown planthopper (SBPH, Laodelphax striatellus). For example, Zhang et al. (2012) identified a cytochrome P450 monooxygenase gene (LsCYP6CW1) that was overexpressed by 22.78-fold in a buprofezin-resistant (59.9-fold) strain of SBPH. Subsequently, Zhang et al. (2017) confirmed by RNA interference (RNAi) approaches that LsCYP6CW1 mediated cross-resistance between buprofezin and pymetrozine in three field populations of SBPH. These studies indicated that cytochrome P450 enzymes were likely involved in buprofezin resistance in SBPH. Nevertheless, despite the extremely high levels of resistance (e.g., > 1000-fold) to buprofezin that were detected in the field populations of BPH, mechanisms of buprofezin resistance, particularly related to possible alterations of buprofezin target site, are poorly understood.
Similar to the mode of action of benzoylphenyureas (BPUs), which belong to Group 15 and function as chitin synthesis inhibitors affecting chitin synthase 1 (Insecticide Resistance Action Committee 2021), buprofezin blocks the incorporation of radiolabeled chitin precursors and prevents formation of a lamellate cuticle to kill insects (De Cock et al. 1990). It has been shown that the I1042M/F mutation on chitin synthase confers very high resistance to BPU, and this mutation can also lead to cross-resistance to buprofezin (Douris et al. 2016). To date, however, this mutation has not been found in BPH or any other hemipteran species which are managed by buprofezin.
The objectives of this research were to: (1) conduct a long-term and large geographical scale-monitoring program for buprofezin resistance in BPH, (2) elucidate molecular mechanism causing extremely high levels of resistance to buprofezin, and (3) validate the resistance mechanism using CRISPR/Cas9 genome editing technology in Drosophila melanogaster model. Our study revealed a novel mutation in chitin synthase 1 gene (chs1) and demonstrated the involvement of the mutation in mediating buprofezin and cyromazine cross-resistance in Drosophila. As cyromazine is considered as an insect molting disruptor that belongs to Group 17 (Insecticide Resistance Action Committee 2021), our results provide new insights into the mode of action of cyromazine which is largely unknown.
Materials and methods
Insects
The susceptible strain (Bup-S) of BPH was initially collected from Hangzhou, Zhejiang Province, in 1995 and has been maintained in the laboratory without exposure to any insecticide since then. The buprofezin-resistant strain (Bup-R) was collected from Haiyan County, Zhejiang Province, in 2007 and has been selected for resistance with buprofezin for more than 10 years. Our study also included several field-collected BPH colonies showing very high resistance to buprofezin. These colonies were collected from different Provinces at different times. All the BPH strains and colonies were reared on rice seedlings under standard conditions of 27 ± 1 °C and 70–80% relative humidity with a 16-h light/8-h dark photoperiod.
The Drosophila line y1 w1118; attP40{nos-Cas9}/CyO was used for genome editing experiments. Line w-;;MKRS/TM6B containing the 3nd chromosome balancers and wild type w1118 were used for outcrossing, balancing, and control experiments. Line w1118;; PBac{RB}kkve03205/TM6B, Tb1 (stock #18,132 at Bloomington Drosophila Stock Center) containing a transposon insertion at the KKV target region was used for complementation test with G932C/TM6B heterozygotes. Flies were cultured at 25 ℃, 60–70% humidity, and 12:12 h photoperiod on standard Drosophila diet.
Insecticide bioassays
The rice-stem dipping bioassay method was used to evaluate the susceptibility of BPH to buprofezin as previously described (Wang et al. 2008c). Briefly, rice plants at the tillering to early booting stage were uprooted, washed thoroughly, cut into an approximately 10-cm-long rice stem with roots, and air-dried. Each insecticide was diluted to a series of concentrations with ultrapure water containing 0.1% Triton-X 100 (Sigma-Aldrich, St. Louis, MO, USA). The prepared rice stems were dipped into appropriate insecticide solution for 30 s and air-dried at room temperature. Rice roots were wrapped with moist cotton wool and put into 500-ml plastic cups. Fifteen third-instar nymphs of BPH were then transferred into each cup with a homemade aspirating device. Each bioassay consisting of 6–10 insecticide concentrations was replicated three times. Ultrapure water only containing 0.1% Triton-X 100 was used as negative controls. All treatments were maintained at 27 ± 1 °C and 70–80% relative humidity with a 16-h light/8-h dark photoperiod. The BPH mortality was assessed at 120 h after the buprofezin treatments.
For insecticide bioassays in Drosophila, we adopted a method using insecticide-incorporated diet. Briefly, fresh Drosophila diet was prepared and cooled to 50 ℃ at room temperature. The diet was then mixed with the pre-prepared insecticide stock solution in a 4:1 volume ratio (ultrapure water was used in controls) and stirred with a magnetic stirrer for at least 1 min, allowing a complete mixing of the diet and insecticides. Subsequently, aliquots of 4-mL toxic diet were added to disposable transparent plastic vials in which the diet solidified. Finally, 15 s-instar larvae were collected and transferred into a plastic vial containing the insecticide-incorporated diet. Each bioassay consisted of 5–6 insecticide concentrations; each was replicated for four to six times. The survivorship of the adult flies was examined after 8–10 d. The bioassay also included controls with no insecticides.
In this study, the detailed information of all insecticides used is as follows: buprofezin (70% WG) was provided by Shaanxi Huarong Kaiwei Biological Co., Ltd. (Shanxi, China); cyromazine (98% TC) was provided by Jiangxi Heyi Chemical Co., Ltd. (Jiangxi, China); hexaflumuron (10% SC) was provided by Dezhou Luba Fine Chemical Co., Ltd. (Shandong, China); chlorfluazuron (5% EC) was provided by Hebei Guanlong Agrochemical Co., Ltd. (Hebei, China); and lufenuron (5% EW) and etoxazole (20% SC) were provided by Shandong Lufeng Pesticide Co., Ltd. (Shandong, China). All the insecticides were dissolved in ultrapure water.
Genetics of buprofezin resistance
The method of genetic analysis was adopted from Roditakis et al. (Roditakis et al. 2016) with minor modifications. Briefly, BPH nymphs of the Bup-S and Bup-R strains were raised to fifth-instar on rice seedlings under the controlled conditions as described above. After the sex was separated based on external morphology, they were raised individually to ensure that the females to be used were virgin. Subsequently, each of 150 virgin females of Bup-R strain was crossed with each of 150 males of Bup-S strain and vice versa. Third-instar nymphs of the F1 generation of reciprocal crosses were subsequently bioassayed with buprofezin to determine LC50 values as described above. The degree of dominance was calculated using the formula D = (2X2–X1–X3)/(X1–X3) (Stone 1968), where X1, X2 and X3 refer to the log (LC50) values of Bup-R strain, F1 generation, and Bup-S strain, respectively. The F1 generation of reciprocal crosses was then back-crossed with the parental Bup-R to check for monogenic resistance. For monogenic recessive inheritance of resistance in reciprocal crosses, a plateau is expected at 50% mortality across a range of discriminating doses (Georghiou 1969).
Sample preparation for bulk segregant analysis
The genetic background of Bup-R was different from Bup-S because Bup-S and BUP-R populations were collected from the field in 1995 and 2007, respectively. However, inbred strains are more conducive in BSA mapping. To obtain inbred Bup-R1 strain, we firstly performed sequential three rounds of mass cross, backcross, and selection using Bup-R and Bup-S. The program is schematically illustrated in Fig. S4. Briefly, 200 males from Bup-R were mass-crossed with 200 virgin females from Bup-S. After 200 males of F1 progeny were backcrossed with 200 virgin females from Bup-S to produce BC1 (RS + SS, 1:1), the BC1 was screened with 30 mg/L buprofezin, which kills the susceptible offspring (SS) and some of the heterozygotes (RS).
Surviving male adults (RS) were backcrossed to Bup-S and screened with buprofezin for six successive generations. Surviving BC7 (RS) were allowed to mate among themselves to generate F2 (RR + RS + SS, 1:2:1). All the F2 progenies were screened with 1000 mg/L buprofezin, which was supposed to kill all SS and RS offspring, leaving only homozygous resistant individuals (RR). Surviving F2 progenies were allowed to continuously mate with Bup-S according to the above steps for two rounds to obtain BUP-R1, which is a near-isogenic resistant strain for Bup-S. After a single virgin Bup-R1 female was crossed to a single Bup-S male, all of F1 progenies were adequately interbred to produce F2 progenies, which were regarded as maximal recombination between the genomes of Bup-R1 and Bup-S strains to break apart haplotypes near resistance mutation. The resulting F2 intercross progenies were then divided into two experimental populations: one was treated with 5 mg/L buprofezin and then the poisoned planthoppers were collected, and the other was treated with 1000 mg/L buprofezin and the vigorous planthoppers were collected. The DNA samples of the two strains after the above screening and the parental lines were extracted for further applications.
Bulk segregant analysis (BSA) genetic mapping
Total genomic DNA was extracted from bulks and at least 3 µg genomic DNA was used to construct paired-end libraries with an insert size of 500 bp using Paired-End DNA Sample Prep kit (Illumina Inc., San Diego, CA, USA). These libraries were sequenced using HiSeq X10 (Illumina Inc., San Diego, CA, USA) NGS platform. Raw reads were then processed to obtain high-quality clean reads according to three stringent filtering standards: 1) removing reads with ≥ 10% unidentified nucleotides (N); 2) removing reads with > 50% bases having Phred quality scores of ≤ 20; and 3) removing reads aligned to the barcode adapter.
To identify single nucleotide polymorphisms(SNPs), filtered reads were aligned to the BPH reference genome (Ma et al. 2021) using Burrows–Wheeler Aligner (BWA, v 0.7.16a-r1181) with parameter ‘mem 4-k 32-M’, where k is the minimum seed length and M is an option used to mark shorter split alignment hits as secondary alignments (Li and Durbin 2009). Variant calling was carried out using GATK UnifiedGenotyper (v3.5). SNPs and InDels were filtered using GATK VariantFiltration function with proper standards (-Window 4, -filter ʹʹQD < 4.0 || FS > 60.0 || MQ < 40.0 ʹʹ, -G_filter ʹʹGQ < 20ʹʹ). All mutations were annotated for genes and function as well as genomic regions using ANNOVAR (Wang et al. 2010).
The SNP-index and the ∆(SNP-Index) of each SNP/Indel were calculated as follows: SNP-index = ADr/(ADd + ADr), ∆(SNP-Index) = SNP-index (recessive)—SNP-index (dominance bulk) (Takagi et al. 2013), where ADr represents recessive allele depth (mutation allele depth in this study) and ADd represents dominance allele depth (wildtype allele depth). Recessive/mutation allele and dominance/wildtype allele in each bulk were polarized according to their grand-parents’ genotypes. We also calculated the statistical confidence intervals of the ∆(SNP-index) under the null hypothesis of no QTLs. For each position, the 99% confidence intervals of the ∆(SNP-index) were obtained following the method described in Takagi et al. (Takagi et al. 2013). Averages of ∆(SNP-index) and SNP-index for each bulk were calculated using a 1,000 kb sliding window with a step size of 10 kb; windows with < 10 SNP/Indel were discarded; windows with ∆(SNP-index) out of 99% confidence intervals were treated as significant windows; overlapped or adjacent significant windows were merged into a large significant genomic region; and the genes in the interval were used as candidate genes.
Extraction of genomic DNA and detection of the mutation frequency
Genomic DNA was isolated individually from Bup-R strain and additional field strains using 350 μl of hot DNA lysis buffer [100 mM Tris, 50 mM ethylenediaminetetraacetic acid (EDTA), 200 mM NaCl, 1% sodium dodecyl sulfate (SDS), pH 8.0]. At least 20 adults from each strain were examined. The genomic DNA of each adult was amplified by PCR using specific primers (Table S4) based on the following protocol: an initial denaturation at 94 ℃ for 2 min; 35 cycles of denaturation at 94 ℃ for 20 s, annealing at 62 ℃ for 30 s, and extension at 72 ℃ for 10 s; and a final extension step at 72 ℃ for 5 min. At the end, all PCR products were directly sequenced by TSINGKE Biotechnology (Beijing, China). Alignment of sequencing results was analyzed using DNAman v.6.0 Software (Lynnon Biosoft, Quebec, Canada).
Genomic engineering strategy
We generated the following mutations including G932C (equivalent to G932C in BPH), I1056M, and both G932C and I1056M simultaneously at the kkv gene in Drosophila by an ad-hoc CRISPR/Cas9 genomic engineering strategy. Potential CRISPR targets in regions of interest were identified using the online tool Optimal Target Finder (Gratz et al. 2014) (http://tools.flycrispr.molbio.wisc.edu/targetFinder/), and three targets with no predicted off-target hits were selected to construct RNA expressing plasmids gRNA936F, gRNA936R/gRNA1056F and gRNA1056R targeting the relevant genomic regions (Fig. S1). The methods of constructing these plasmids were described previously (Douris et al. 2016). We constructed de novo three donor plasmids for Homology-Directed Repair (HDR) (GenScript Biotech, Piscataway, NJ, USA), each containing 2 ~ 800 bp homology arms flanking the relevant CRISPR target region, with certain modifications compared to wild-type genomic sequence (Fig. S1).
Generation and screening of genome-modified Drosophila
We first identified the BPH orthologous gene chs1 in Drosophila (known as krotzkopf verkehrt or kkv) and checked the nucleotide sequence of an 891-bp fragment of kkv exon 6 (corresponding to 3R: 5,381,808: 5,382,698 at the BDGP6 genome assembly). To generate various mutations in kkv including the G932C corresponding to G932C in BPH, I1056M corresponding to I1042M in P. xylostella, and both G932C and I1056M mutations, we injected y1w1118; attP40{nos-Cas9}/CyO (Gokcezade et al. 2014) embryos with the respective gRNAs/donor plasmid mixes and screened progeny for genome-modified allele. The presences of HDR derived alleles were 12 out of 24 different lines that gave G1 progeny for 932C mutation, 10 out of 24 for 1056 M, and 6 out of 24 for both 932C and 1056 M (Table S4).
Subsequently, the homologous repaired individuals are purified through successive generations of hybridization with w1118 and balancer lines (supplementary information). Several independent lines were established and at least one became readily homozygous after balancing for the 1056 M. All of these homozygous mutation progenies were normal in the larval stage, whereas none of the all-independent lines in the adult stage can survive normally and gave homozygous progeny for the 932C mutation and both 932C and I1056M mutations. All lines were verified by sequencing the relevant genomic region and shown to be genome-modified as expected, which beard the correct mutations and other inserted markers in the kkv gene. Detailed information for injection of gRNAs/donor, genome modification strategy, positive rates of the originals, crossing schemes for genome-modified flies, is provided in Fig. S1, Fig. S2, and Table S5.
Analysis of fitness cost in genome-modified Drosophila
A total of four Drosophila lines (w1118, w-;;932C/TM6B, w-;;(932C/I1056M)/TM6B, w-;;1056 M) and 40 adult females and 40 adult males with full copulation in yeast-containing foods for 3 days for each line were placed in 9-cm diameter plastic Petri dishes with cherry-agar plates layered with yeast. After the flies were adapted for several hours, the plates were replaced to allow the flies to lay their eggs for ∼2 h followed by removing all the adults from the plates. Then, 20 newly hatched first-instar larvae were immediately transferred to the small vials with fly diet (four replicates for w1118 and w-;;1056 M, six replicates for w-;;932C/TM6B, w-;;(932C/I1056M)/TM6B) and allowed them to grow under the standard conditions (25 ± 1 °C, relative humidity 70 ± 10%, LD 12:12). All larval and pupal developmental times were recorded for w1118 and w-;;1056 M, and only larval and pupal developmental times with a normal phenotype were recorded for w-;;932C/TM6B and w-;;(932C/1056 M)/TM6B.
Analysis of fitness cost in BPH
Bup-R and Bup-S strains were maintained in an incubator under standard rearing conditions as described above. To determine the developmental time from egg to adult, 200-pairs of fully mated adults of each strain were placed in a 1,000-ml glass beaker containing rice seedlings for oviposition. The females were allowed to lay eggs for 12 h and removed afterward. After about 7 days, 100 newly hatched first nymphs were individually transferred to 100 disposable transparent plastic vials containing rice seedings. Each molting time was recorded every 12 h until adult emergence and the emergence rates were calculated. After the emergence, the survivorship was recorded every 12 h until the adult died.
Spontaneous locomotion assay in Drosophila
Within 12 h after emergence, individual flies from each of the four Drosophila lines were transferred into the round wells (2-cm diameter and 3-mm height) covered by regular food in a recoding chamber, and their locomotion was recorded for 2 h. The average walking velocity during the 2-h recording was quantified using the ZebraLab software system (ViewPoint Life Sciences, Montreal, Quebec, Canada) as previously described (Wu et al. 2019). Likewise, detailed motion videos of the four lines were recorded by a camera.
Investigation of potential factors affecting fitness of the G932C allele in Drosophila.
To investigate if the inability to produce homozygous adults bearing the G932C allele was due to a potential lethality of the specific allele in Drosophila or to other factors genetically linked to kkv at the third chromosome, we carried out additional studies as previously described (Douris et al. 2017) with some modifications. Briefly, heterozygous w1118; PBac(RB)kkve03205/TM6B, Tb1 (stock #18,132 at Bloomington Drosophila Stock Center, Bloomington, IN, USA) containing a transposon insertion at the kkv target region was used for complementation test with 932C/TM6B heterozygotes. Our result indicated that all the progenies normally growing in the adult stage from all crosses beared the TM6B balancer chromosome; thus, no 932C/18132 complementation is apparently viable, which confirmed that the observed lethality was linked to the corresponding genomic region 3R:5,378,093; 3R:5,392,866 containing the target kkv region, presumably due to the 932C mutation itself.
Statistical analysis
The median lethal concentrations (LC50) and their 95% fiducial limits (FL) were estimated using POLO-plus program (Version 2.0) (LeOra Software, Petaluma, CA, USA) for BPH. If the 95% FLs of two LC50 values do not overlap, the two LC50 values were considered to be significantly different. The resistance ratio (RR) was calculated by dividing the LC50 value of a resistant strain by that of the susceptible strain. Insecticide resistance of the field populations was classified as: RR < fivefold as susceptible, RR = 5–tenfold as low resistance, RR = 10–100-fold as medium resistance, and RR > 100-fold as high resistance (Mu et al. 2016). Data from the fitness cost analysis in genome-modified Drosophila were compared for all the four lines using one-way ANOVA followed by Tukey HSD test, whereas data from the fitness cost analysis were compared between the BPH strains using Student’s t-test.
Results
Monitoring buprofezin resistance in BPH field populations
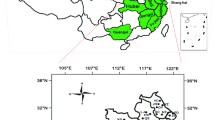
Our long-term (25 years from 1996 to 2020) and large geographical scale (11 provinces and cities in China) resistance monitoring program for buprofezin in 179 BPH field populations revealed high (40- to 160-fold) to extremely high (> 160-fold) levels of buprofezin resistance in 2013 and thereafter (Fig. 1A, B). From 2015 to 2020, more than 1,000 and up to 5,622.3 fold resistance levels were detected in the most field populations examined (Fig. 1B).
(A) A total of 179 populations from 11 provinces and cities of China were collected during 1996–2020. The map was generated by software Adobe Photoshop CS5 version (San Jose, CA, USA, http://www.adobe.com/products/photoshop.html) based on our data. (B) Resistance ratios to buprofezin of all collected BPH populations were determined from 1996 to 2020. Some of these data were derived from our previous publications (Wang et al. 2008c, 2008d; Wu et al. 2018).
Genetic basis of buprofezin resistance
To investigate genetic basis of buprofezin resistance in BPH, we further selected a buprofezin-resistant strain (Bup-R) derived from a field population (HY2007). In comparison with a laboratory buprofezin-susceptible strain (Bup-S), Bup-R showed extremely high level of buprofezin resistance with a ratio of > 10,000-fold (Fig. 2 and Table S1). The degree of dominance analysis in F1 progenies of reciprocal crosses revealed that buprofezin resistance was inherited autosomal incompletely recessive mode of inheritance and not maternally inherited or sex-linked. After female F1 hybrids of (Bup-R)♀ × (Bup-S)♂ crosses were backcrossed with males of Bup-R and tested for monogenic resistance, the obtained experimental dose–response curve for buprofezin had a plateau at 50% mortality, indicating buprofezin resistance segregates as a single resistance locus (X2 = 22.03, df = 6, P < 0.05) (Fig. 2).
Concentration response relationship between buprofezin and BPH strains including Bup-S, Bup-R, reciprocal crosses of F1 hybrids: Bup-R(♂) X Bup-S(♀) and F1’ hybrids: Bup-R(♀) X Bup-S(♂). High resistance is inherited recessively and is not maternal [as compared reciprocal F1s (triangles) with parental strains (circles)]. The mortality plateau at 50% for F1 X Bup-R backcross revealed that resistance was controlled by a single major factor.
Identification of the buprofezin resistance locus
We used BSA genetic mapping method to identify the resistance locus in Bup-R. Five candidate regions associated with the resistance traits were located on chromosome 3 (Fig. 3A). We further selected the candidate range with the limitation of non-synonymous mutations on exons, based on the possible target site hypothesis, as indicated by the striking resistance phenotype. The causal region for recessive resistance in Bup-R1 is about 13.33 Mb in length, which harbors 38 genes with 63 non-synonymous SNPs (Fig. 3B). A chitin synthase gene was found in this region (Nlug06525), whereas a non-synonymous substitution from glycine (G) to cysteine (C) at position 932 (G932C) was identified to the fifteenth exon of Nlug06525 (Fig. 3C). The remaining 37 genes (Table S2) are not involved in either chitin biosynthesis or transport (Candy and Kilby 1962).
(A) The results from BSA genetic scans for buprofezin resistance by average Δ (SNP-index) graph based on the data of Bup-S pool and Bup-R pool against reference genome (Ma et al. 2020) are shown. The peak of target region was shown on Chromosome 3 including five regions (12.04 M-13.54 M, 22.26–35.59 M, 39.54–41.77 M, 48.72–53.13, 84.81–86.86 M). The confidence limits were revealed with red lines (P < 0.01). (B) Schematic diagram of predicted gene affecting buprofezin resistance in minimal candidate region. We narrowed the candidate interval using a limitation condition with non-synonymous mutation on the exon. A total of 36 genes were screened out, of which only one gene was related to the chitin synthetic pathway. (C) Diagram of the gene structure of a candidate gene with a non-synonymous mutation on exon 15 leading to an amino acid substitution.
A chs1 mutation G932C associated with buprofezin resistance
To verify whether the G932C substitution mutation in chs1 revealed by BSA was at the same site as the previously reported mutation in other arthropod species, we aligned amino acid sequences deduced from the chs1 orthologous genes from BPH, diamondback moth (Plutella xylostella) and two-spotted spider mite (Tetranychus urticae). We found that the previously identified mutations of I1017F in T. urticae and I1042M in P. xylostella corresponded to a position of 1052 in BPH chs1 (Fig. 4A). To further determine whether there were variants related to buprofezin resistance at locus 1052 of chs1 in BPH, we subsequently sequenced the full-length chs1 cDNAs from both Bup-S and Bup-R strains of BPH, but did not find any mutation at position 1052 of chs1 in BPH. We only identified the G932C mutation near the 3’ end which corresponds to the C-terminal region of the chitin synthase 1 amino acid sequence in Bup-R strain (Fig. 4A and B), but this mutation was completely absent in Bup-S strain (Table S2).
To determine whether the G932C mutation was associated with buprofezin resistance, we examined the mutation frequency in several field-collected BPH populations showing different levels of buprofezin resistance. Our results showed that there was a positive correlation between the resistance ratio and the G932C mutation frequency (Fig. 4C, R2 = 0.76 (F = 15.82, df = 5, P = 0.01) and Table S3). Further selection of a field-collected colony (YC2017) with buprofezin at the median lethal concentration (LC50) significantly increased the frequency of the 932C mutation in the survivors.
(A) Top: Schematic representation of domain architecture of chitin synthase 1 redrafted based on (Douris et al. 2016). 5TMS, five transmembrane spans; CC, coiled-coil motif; CD, catalytic domain; NTR, N-terminal region; CTR, C-terminal region. Cylindrical shells represent transmembrane domains. Arrows point to signature sequences QRRRW (catalytic domain) and WGTR (N-terminal region). Bottom: Aligned amino acid sequences of helix1 and helix 5 in the 5TMS clusters of chitin synthase 1 of T. urticae (Tu; S, etoxazole susceptible; R, etoxazole resistant), N. lugens (Nl; S, buprofezin susceptible; R, buprofezin resistant), P. xylostella (Px; S, benzoylureas susceptible; R, benzoylurea resistant), D. melanogaster (Dm) and T. castaneum (Tc). The position of the G932C substitution in buprofezin resistant N. lugens and I1056M/F substitution in benzoylureas resistant P. xylostella (I1017F in etoxazole-resistant mites) is indicated in gray. (B) Chromatograms of the nucleotide sequences of the mutation site of Nlchs. (C) Correlation between the ratio of buprofezin resistance and the frequency of the G932C mutation of Nlchs. Linear regressions are shown for significant correlations (df = 5, P = 0.01). P values are shown for Spearman Rank Order correlations. Underlying data can be found in S1 Data.
Generation and fitness analysis of CRISPR/Cas9 genome-modified Drosophila
As previously described (Douris et al. 2016), we identified the orthologous of BPH chs1 in Drosophila and injected the three corresponding gRNAs/donor plasmid mixes into y1w1118; attP40{nos-Cas9}/CyO (Gokcezade et al. 2014) embryos, including G932C corresponding to G932C in BPH, I1056M corresponding to I1042M in P. xylostella, and the G932C and I1056M combination (double mutant) [see Supplemental Information (SI) Text for details]. The three genome-modified Drosophila lines were verified by sequencing the relevant genomic region.
We firstly investigated the effect of amino acid substitution on the fitness of Drosophila by comparing the differences in developmental time, pupation rate, eclosion rate, and adult locomotion among the four lines including a control w1118 line and three genome-modified lines (932C, 1056 M, and 932C/1056 M). No significant difference was observed for developmental time and pupation rate (Fig. 5A. and B). However, two Drosophila lines bearing the 932C mutation (i.e., 932C and 932C/1056 M) showed a significant decrease in eclosion rate compared with the control w1118 line (Fig. 5C). Specifically, some Drosophila with the 932C mutation were unable to completely get rid of the pupal cuticle during their eclosion, which led to mortality (Fig. 5D).
Although other Drosophila flies bearing the 932C mutation can eclose successfully, their locomotion was significantly impaired and they cannot move normally, resulting in reproductive failure (Fig. 5E and Video 1). We further ruled out other factors that might potentially lead to a significant fitness cost in the 932C mutant flies by hybridizing with the chs1 mutated flies (Douris et al. 2017) (see SI Text for details). Additionally, we investigated certain life table parameters in BPH for Bup-S and Bup-R strains. No significant difference was found for nymphal development time, emergence rate and fecundity between Bup-S and Bup-R strains (Fig. S3).
(A and B) Developmental time in larval stage, pupal stage and pupation rate are compared for different Drosophila lines used in this study (n ≥ 30 for each line). There is no significant difference. (C) The eclosion rates are compared for different Drosophila lines used in this study. Drosophila bearing the 932C mutation, including lines 932C/1056 M, have a significantly lower emergence rate as compared with lines 1056 M and w1118 (n ≥ 15 for each repetition, total five repetitions in each line). (D) A phenotypic map of Drosophila bearing 932C mutation that could not completely get rid of the pupal cuticle. (E) shows the locomotor capacity of adults after eclosion of different strains. The locomotor capacity of two 932C-bearing mutation lines was seriously affected. These results suggest that the 932G mutation has significant negative effects on Drosophila emergence and coordination of adult movement (n = 12 for each line). The variance analysis of all data is assessed by one-way ANOVA with Tukey HSD test. The bars of the means represent SD (*, p < 0.05; ***, p < 0.001; ****, p < 0.0001; ns, no significant).
Video 1. Record of motor ability for four Drosophila lines
The video shows significant effects of the buprofezin resistance mutation on the motor ability of the flies.
Drosophila bearing the 932c mutation exhibits resistance to buprofezin and cyromazine
Firstly, we tested the susceptibility of four Drosophila lines (w1118, 932C, 1056 M, and 932C/1056 M) to etoxazole and five BPU insecticides (chlorbenzuron, hexaflumuron, chlorfluazuron, lufenuron, and diflubenzuron). Our results showed that the two Drosophila lines bearing the 1056 M mutation (i.e., 1056 M and 932C/1056 M) were highly resistant to etoxazole and all the five BPUs (Table 1). In contrast, the Drosophila line bearing the 932C mutation was not resistant to etoxazole or any of the five BPUs (Table 1). Although the two Drosophila lines bearing the 1056 M mutations showed a marginal resistance to buprofezin (4.5-fold for 1056 M mutant and 2.6-fold in 932C/1056 M) as compared with the control w1118 line, the most pronounced buprofezin resistance (8.3-fold) was found in the line bearing the 932C mutation (Table 1). Furthermore, this mutation also conferred a low level (3.2-fold) of cross-resistance to cyromazine.
Discussion
Since buprofezin was introduced to control BPH in China in 1985, and extensive applications of this insecticide have led to the development of high resistance in BPH populations (Fig. 1B). Indeed, a vast majority of the BPH field populations examined in our study were highly resistant to buprofezin (> 100-fold) in 2013. Within only 2 years after 2013 (i.e., 2015), extremely high resistance (> 1,000-fold) was detected in the most field populations examined. In 2020, the highest resistance level detected was 5,622.3-fold in Xiantao City, Hubei Province. Such high levels of resistance to buprofezin allowed us to conduct some detailed studies to reveal resistance mechanisms in BPH.
To understand molecular mechanisms of buprofezin resistance in BPH, we firstly identified a Bup-R population harboring recessive, monogenic resistance to buprofezin. Subsequently, we applied BSA-based genetic mapping method and revealed a novel non-synonymous amino acid substitution mutation (G932C) in chs1 which was located on chromosome 3 (Fig. 3A). Surprisingly, the presence and frequency of the G932C mutation were highly correlated with buprofezin resistance in BPH (Fig. 4C). Our discovery of the same mutation of G932C in buprofezin resistant SBPH (data not shown) further supports our notion that the G932C mutation plays a significant role in buprofezin resistance.
Our findings were further validated using Drosophila model. Introduction of G932C mutation in Drosophila by CRISPR/Cas9 coupled with HDR genome modification approach showed significant resistance to buprofezin and cyromazine, but not BPUs and etoxazole. These results indicate that, similar to BPUs and etoxazole, buprofezin also targets chitin biosynthesis to confer its insecticidal activity (Douris et al. 2016), but the main binding sites of buprofezin appears to be different from those of BPUs and etoxazole. In addition, our results also provide a compelling evidence that cyromazine, which is considered as an insect molting disruptor (Insecticide Resistance Action Committee 2021), may also interfere chitin biosynthesis.
BSA is a quantitative trait locus (QTL) mapping technique for identifying genomic regions containing genetic loci affecting a trait of interest (Michelmore et al. 1991). The biggest advantage of BSA over ʹʹregularʹʹ QTL analysis is that there is no need for genotyping and phenotyping each of hundreds of individuals in a segregating population (Chu et al. 2016). However, it requires genomic resources and homozygous samples exhibiting extremely different traits. For the first time, Leeuwen et al. (2012) developed a population-level bulk segregant mapping method based on high-throughput genome sequencing to identify a locus for resistance to etoxazole in the two-spotted spider mite. Afterward, several studies applying BSA to localize resistance loci to a small genomic region have been reported (Demaeght et al. 2014; Fotoukkiaii et al. 2021). By using the same approach, we successfully localized the buprofezin resistance loci on chs1 gene encoding chitin synthase 1 in BPH.
Chitin synthases are highly complex proteins that have never been heterologously expressed and their structure is largely unknown in insects (Zhu et al. 2016). To validate the contribution of the G932C mutation to buprofezin resistance in BPH, we used a genome editing approach by generating its corresponding mutation (i.e., G932C) in Drosophila. Although only 8.3-fold resistance to buprofezin was observed in genome-modified Drosophila bearing the G932C mutation, these results provided additional support to our hypothesis that this mutation contributed to high levels of resistance to buprofezin in BPH. The low levels of resistance in the genome-modified Drosophila could be due to the species difference and/or the absence of other resistance mechanisms against buprofezin in Drosophila.
Insect chitin synthase contains approximately 15 transmembrane helices with flanking catalytic domains located on the cytoplasmic side of the plasma membrane (Zhu et al. 2016). Previous studies suggested that the transmembrane helices located at the C-terminal region of the enzyme are involved in pore formation, which is required for the translocation of nascent chitin polymers across the membrane (Merzendorfer 2006). This information allowed us to identify the position of the G932C mutation in chitin synthase 1 gene of BPH. Clearly, the 932C mutation is located at the first one of the five transmembrane spans (5-TMS) after the catalytic domain, which suggests that buprofezin may bind to the TMS. Specifically, the sulfhydryl group of the cysteine residue in the first TMS of chitin synthase 1 in the buprofezin-resistant BPH instead of the hydrogen of the glycine residue in the susceptible BPH is likely to change spatial conformation of chitin synthase 1 as glycine is the smallest amino acid residue. Such a change could interfere with the interactions between buprofezin and chitin synthase 1, and therefore consequently reduce BPH’s susceptibility to buprofezin.
Our study also showed a significant fitness cost associated with the G932C mutation in genome-modified Drosophila, whereas no fitness cost was associated with the same mutation in BPH (Fig. 5 and Fig. S3). This phenomenon is likely to be similar to that of G4946E mutation found in ryanodine receptor (RyR) gene. Previous studies have shown that the G4946E mutation was closely associated with diamide insecticide resistance in lepidopteran pests, but the same mutation that was introduced to Drosophila resulted in a lethal phenotype (Douris et al. 2017). Nevertheless, the exact mechanism leading to differential fitness cost between BPH and Drosophila bearing the same mutation remains to be determined.
In summary, our results have provided multiple lines of evidences to support our conclusion that the G932C point mutation of chs1 confers high levels of resistance to buprofezin in BPH, which include: 1) Our genetic analysis of resistance confirmed that the resistance to buprofezin in Bup-R was controlled by a single gene; 2) by using BSA approaches to localize the resistance locus in Bup-R1, we found only one gene (chs1) that is involved in chitin biosynthesis (Candy and Kilby 1962) within the causal region for buprofezin resistance; 3) the gene chs1 encoding chitin synthase 1, a known target of buprofezin, beared the non-synonymous mutation (G932C) and the mutation frequency was highly correlated with the levels of buprofezin resistance in BPH; 4) our new inbred Bup-R1 strain created from three rounds of sequential mass cross, backcross, and selection from the Bup-R and Bup-S strains showed about 94% of its genetic background with Bup-S and > 1000-fold resistance to buprofezin; and 5) introduction of G932C mutation in Drosophila by CRISPR/Cas9 coupled with HDR genome modification approach led to buprofezin resistance.
Furthermore, our studies have provided new insights into the mode of action of cyromazine. To date, although there have been some toxicological studies of cyromazine, its specific mode of action is still unknown. Miller et al. (Miller et al. 1981) proposed that cyromazine might act on the process of chitin synthesis or interact with certain epidermal proteins in insects (Binnington 1985; Miller et al. 1981). In housefly, it was speculated that cyromazine might act on dihydrofolate reductase (DHFR), but several subsequent experiments proved that cyromazine can only slightly inhibit DHFR activity, indicating that the inhibition of DHFR is unlikely a primary mode of action for cyromazine (Bel et al. 2000; El-Oshar et al. 1985). In Drosophila, it has been documented that an orthologous gene (CG32743) of Smg1 of worms and mammals, which encodes phosphatidylinositol kinase-like kinase, has cyromazine resistance alleles (Chen et al. 2006). Because human Smg1 has been shown to have a role in DNA damage pathways, it has been suggested that cyromazine might interfere with nucleic acid metabolism. In our study, however, we found that the G932C mutation of chs1 not only contributed to buprofezin resistance in BPH but also led to resistance to cyromazine in the genome-modified Drosophila. All these results suggest that cyromazine may attack chitin biosynthesis in these insect species.
In conclusion, our study showed for the first time that the novel G932C mutation of chs1 contributed to high levels of resistance to buprofezin in BPH. This finding was strongly supported by the results from our confirmative experiments in both buprofezin-resistant BPH and genome-modified Drosophila. However, the chs1 mutation conferring buprofezin resistance was different from those conferring BPU and etoxazole resistance in arthropods. Our results also implicated that cyromazine might target chitin biosynthesis as demonstrated by the cross-resistance between buprofezin and cyromazine induced by the same mutation in Drosophila model. This finding may direct new research for better understanding of cyromazine’s mode of action in arthropods.
Author contribution
BZ, SFW, and CFG designed the experiments. BZ, FRC, YTL, DG, YJZ,ZRF, and LXW performed the experiments and the data analysis. BZ, JV, KYZ, SFW and CFG wrote the manuscript. All authors read and approved the final manuscript.
References
Bel Y, Wiesner P, Kayser H (2000) Candidate target mechanisms of the growth inhibitor cyromazine: Studies of phenylalanine hydroxylase, puparial amino acids, and dihydrofolate reductase in dipteran insects Archives of insect biochemistry and physiology 45:69–78
Binnington K (1985) Ultrastructural changes in the cuticle of the sheep blowfly. Lucilia, Induced by Certain Insecticides and Biological Inhibitors Tissue and Cell 17:131–140
Candy D, Kilby B (1962) Studies on chitin synthesis in the desert locust. J Exp Biol 39:129–140
Chen Z, Robin C, Damiano J et al (2006) Positional cloning of a cyromazine resistance gene in Drosophila melanogaster. Insect Mol Biol 15:181–186. https://doi.org/10.1111/j.1365-2583.2006.00622.x
Chu Y, Clevenger J, Hovav R et al. (2016) Chapter 7 - Application of Genomic, Transcriptomic, and Metabolomic Technologies in Arachis Species. In: Stalker HT, F. Wilson R (eds) Peanuts. AOCS Press, pp 209–240. doi:https://doi.org/10.1016/B978-1-63067-038-2.00007-1
De Cock A, Ishaaya I, Degheele D et al. (1990) Vapor toxicity and concentration-dependent persistence of buprofezin applied to cotton foliage for controlling the sweetpotato whitefly (Homoptera: Aleyrodidae) J Econ Entomol 83:1254–1260
Demaeght P, Ukken, FP, Rubinstein, CD et al. (2014) High resolution genetic mapping uncovers chitin synthase-1 as the target-site ofthe structurally diverse mite growth inhibitors clofentezine, hexythiazox and etoxazole in Tetranychus urticae Insect Biochem Mol Biol 51:52–61
Douris V, Papapostolou, KM, Ilias A et al. (2017) Investigation of the contribution of RyR target-site mutations in diamide resistance by CRISPR/Cas9 genome modification in Drosophila Insect Biochem Mol Biol 87:127–135
Douris V, Steinbach D, Panteleri R et al (2016) Resistance mutation conserved between insects and mites unravels the benzoylurea insecticide mode of action on chitin biosynthesis. Proc Natl Acad Sci U S A 113:14692. https://doi.org/10.1073/pnas.1618258113
El-Oshar M, Motoyama N, Hughes P et al. (1985) Studies on cyromazine in the house fly, Musca domestica (Diptera: Muscidae) J Econ Entomol 78:1203–1207
Fotoukkiaii SM, Wybouw N, Kurlovs AH et al. (2021) High-resolution genetic mapping reveals cis-regulatory and copy number variation in loci associated with cytochrome P450-mediated detoxification in a generalist arthropod pest PLoS genet 17:e1009422
Georghiou GP (1969) Parasitological review. Genetics of Resistance to Insecticides in Houseflies and Mosquitoes Exp Parasitol 26:224–255. https://doi.org/10.1016/0014-4894(69)90116-7
Gokcezade J, Sienski G, Duchek P (2014) Efficient CRISPR/Cas9 Plasmids for Rapid and Versatile Genome Editing in Drosophila G3-Genes Genomes Genetics 4:2279–2282 doi:https://doi.org/10.1534/g3.114.014126
Gratz SJ, Ukken FP, Rubinstein CD et al. (2014) Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila Genetics 196:961–971 doi:https://doi.org/10.1534/genetics.113.160713
Khush GS (1999) Green revolution: preparing for the 21st century Genome 42:646–655
Li H, Durbin R (2009) Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform Bioinformatics 25:1754–1760
Ma W et al (2021) Chromosomal-Level Genomes of Three Rice Planthoppers Provide New Insights into Sex Chromosome Evolution Mol Ecol Resour 21:226–237. https://doi.org/10.1111/1755-0998.13242
Merzendorfer H (2006) Insect chitin synthases: a review. J Comp Physiol B Biochem Syst Environ Physiol 176:1–15. https://doi.org/10.1007/s00360-005-0005-3
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A 88:9828–9832. https://doi.org/10.1073/pnas.88.21.9828
Miller R, Corley C, Cohen C et al. (1981) CGA 19255 and CGA 72662: mode of action and efficacy against flies in the laboratory and when administered to cattle as feed additive Southwest Entomol 6:272–278
Mu XC, Zhang W, Wang LX et al (2016) Resistance monitoring and cross-resistance patterns of three rice planthoppers, Nilaparvata lugens, Sogatella furcifera and Laodelphax striatellus to dinotefuran in China. Pestic Biochem Physiol 134:8–13. https://doi.org/10.1016/j.pestbp.2016.05.004
Pener MP, Dhadialla TS (2012) Chapter One - An Overview of Insect Growth Disruptors; Applied Aspects. In: Dhadialla TS (ed) Advances in Insect Physiology, vol 43. Academic Press, pp 1–162. doi:https://doi.org/10.1016/B978-0-12-391500-9.00001-2
Roditakis E, Steinbach D, Moritz G et al. (2016) Ryanodine receptor point mutations confer diamide insecticide resistance in tomato leafminer, Tuta absoluta (Lepidoptera: Gelechiidae) Insect Biochem Mol Biol 80:11–20
Stone B (1968) A formula for determining degree of dominance in cases of monofactorial inheritance of resistance to chemicals. Bull World Health Organ 38:325
Takagi H, Abe A, Yoshida K et al. (2013) QTL‐seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations The Plant Journal 74:174–183
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data Nucleic Acids Res 38:e164-e164
Wang L, Fang J, Liu B (2008a) Relative toxicity of insecticides to Laodelphax striatellus (Fallén)(Homoptera: Delphacidae) and the resistance of field populations from different areas of East China Acta Entomologica Sinica 51:930–937
Wang Y, Chen J, Zhu YC et al. (2008b) Susceptibility to neonicotinoids and risk of resistance development in the brown planthopper, Nilaparvata lugens (Stål)(Homoptera: Delphacidae) Pest Manag Sci 64:1278–1284
Wang Y, Gao C, Xu Z et al. (2008c) Buprofezin susceptibility survey, resistance selection and preliminary determination of the resistance mechanism in Nilaparvata lugens (Homoptera: Delphacidae) Pest Manag Sci 64:1050–1056 doi:https://doi.org/10.1002/ps.1606
Wang Y, Gao C, Zhu Y et al. (2008d) Imidacloprid susceptibility survey and selection risk assessment in field populations of Nilaparvata lugens (Homoptera: Delphacidae) J Econ Entomol 101:515–522
Wu S, Guo C, Zhao H et al. (2019) Drosulfakinin signaling in fruitless circuitry antagonizes P1 neurons to regulate sexual arousal in Drosophila Nat Commun 10:4770 doi:https://doi.org/10.1038/s41467-019-12758-6
Wu S, Zeng B, Zheng C et al. (2018) The evolution of insecticide resistance in the brown planthopper (Nilaparvata lugens Stål) of China in the period 2012–2016 Sci Rep 8:4586 doi:https://doi.org/10.1038/s41598-018-22906-5
Zhu KY, Merzendorfer H, Zhang W, e al. (2016) Biosynthesis, Turnover, and Functions of Chitin in Insects. Annu Rev Entomol 61:177–196
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 31972298 & 32022011) and the Jiangsu Agriculture Science and Technology Innovation Fund (CX [19]3003). We thank Plant Protection Stations of Shanghai City, Jiangsu Province, Anhui Province, Zhejiang Province, Hubei Province, Jiangxi Province, Fujian Province, Guangdong Province, Guangxi Province and Hainan Province for helping in collecting tested populations of N. lugens
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors have no competing interest in this work.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Emmanouil Roditakis .
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zeng, B., Chen, FR., Liu, YT. et al. A chitin synthase mutation confers widespread resistance to buprofezin, a chitin synthesis inhibitor, in the brown planthopper, Nilaparvata lugens. J Pest Sci 96, 819–832 (2023). https://doi.org/10.1007/s10340-022-01538-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-022-01538-9