Abstract
Tiancimycin (TNM) A, a recently discovered enediyne natural product from Streptomyces sp. CB03234, showed rapid and complete killing of cancer cells and could be used as a payload in antibody drug conjugates. The low yield of TNM A in the wild-type strain promoted us to use ribosome engineering and fermentation optimization for its yield improvement. The Streptomyces sp. CB03234-R-16 mutant strain with a L422P mutation in RpoB, the RNA polymerase β-subunit, was obtained from the rifamycin-resistant screening. After fermentation optimization, the titers of TNM A in Streptomyces sp. CB03234-R-16 reached to 22.5 ± 3.1 mg L−1 in shaking flasks, and 13 ± 1 mg L−1 in 15 L fermentors, which were at least 40-fold higher than that in the wild-type strain (~ 0.3 mg L−1). Quantitative real-time RT-PCR revealed markedly enhanced expression of key genes encoding TNM A biosynthetic enzymes and regulators in Streptomyces sp. CB03234-R-16. Our study should greatly facilitate the future efforts to develop TNM A into a clinical anticancer drug.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
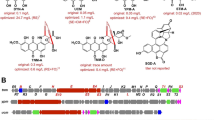
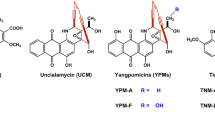
Enediyne natural products, characterized by the conjugation of a double bond with two acetylenic groups within a 9- or 10-membered macrocycle, are among the most cytotoxic natural products to date and several of them have been developed into clinical anticancer drugs (Fig. 1) [23, 34, 35, 37]. Among a dozen of enediynes discovered, the antitumor enediyne chromoprotein neocarzinostatin has been used for hepatoma treatment in Japan since 1994, while the chromoprotein C-1027 was reported to undergo phase II clinical trials in China [19, 32]. Most recently, gemtuzumab ozogamicin (GO), an anti-CD33 antibody–calicheamicin conjugate, was reapproved for acute myeloid leukemia (AML), and inotuzumab ozogamicin, an anti-CD22 antibody–calicheamicin conjugate, was approved for acute lymphoblastic leukemia [18, 31]. GO was actually the first antibody drug conjugates (ADC) on the markets, and the only AML drug to improve patient survival in the last three decades, despite its volunteering withdraw in 2010 [2, 8]. Therefore, highly potent payloads, such as enediynes, are promising drug leads to be used as warheads for ADC drugs [36]. Tiancimycin (TNM) A, a 10-membered enediyne natural product from Streptomyces sp. CB03234, was discovered through a novel genome mining approach (Fig. 1). It was the only new enediyne natural product discovered in the past decade and showed strong inhibitory activities against various tumor cell lines [7, 44]. Remarkably, TNM A exhibited rapid and complete killing of cancer cells, in contrast to the payloads of Adcetris and Kadcyla, two ADC drugs currently on the markets. Therefore, it has very high potential to be developed as effective anticancer drugs.
The Streptomyces sp. CB03234 wild-type strain produces TNM A in a low titer, with only 1.2 mg purified from 6-L fermentation [44]. The low titer of TNM A was similar to the initial titers of the known anthraquinone enediynes, such as dynemicin A and uncialamycin (Table S1). For example, dynemicin A was first isolated with an estimated yield of 0.1 mg L−1, and the production of dynemicin A in 10,000 L was reported to have a titer of 1.2–1.5 mg L−1, which was subsequently optimized to 24.7 mg L−1 in shaking flasks [16, 17]. Uncialamycin was produced from Streptomyces uncialis, with ~ 0.3 mg isolated from 16 L fermentation on solid agar medium [7]. Since the direct access to these fascinating compounds through total synthesis remains impractical, the low titers significantly limit their clinical development [21, 22].
There are many effective strategies to improve the production of secondary metabolites in microorganisms [1, 3, 6, 24, 27, 30, 41]. Many factors would contribute to the titer or the production rate for the targeted secondary metabolites, including the metabolic potential of the producers, and fermentation factors, such as nutrient supply, dissolved oxygen (DO), temperature, and pH. Ribosome engineering is a cost-effective, rapid and a rational approach to obtain mutants with improved titers for secondary metabolite production [25, 26]. By screening for antibiotic resistance targeting bacterial ribosome or RNA polymerase, isolated mutants could have hyperphosphorylated guanosine nucleotide (ppGpp) synthesis activities to induce the stringent response, resulting in extensively reprogramming of the cellular metabolic patterns and even the overexpression of certain pathway specific transcriptional regulators [4, 40].
In this report, we utilized a combination of ribosome engineering and fermentation optimization strategy to rapidly improve the TNM A titer from ~ 0.3 mg L−1 in the Streptomyces sp. CB03234 wild-type strain to 22.5 ± 3.1 mg L−1 (in shaking flasks) and 13.0 ± 1.0 mg L−1 (in 15-L fermenters) in the Streptomyces sp. CB03234-R-16 mutant strain. Quantitative real-time PCR revealed markedly enhanced expression of key genes encoding enzymes and regulators for TNM A biosynthesis in the mutant, in comparison to the wild-type. The current titer of TNM A is comparable to those from industrial enediyne producing strains, such as dynemicin A (24.7 mg L−1) [16] or esperamycin (8.1 mg L−1) [15]. Our findings would greatly facilitate the future efforts to develop TNM A into a clinical anticancer drug.
Materials and methods
Bacterial strains, culture conditions and general methods
All chemical and biological regents, such as antibiotics, inorganic salts, carbon and nitrogen sources used in this study were from common commercial sources, unless otherwise specified. The Streptomyces sp. CB03234 wild-type and mutant strains were grown at 30 °C on ISP4 for sporulation. The Streptomyces sp. CB03234-R-16 mutant strain was deposited at China Center for Type Culture Collection under accession No. CCTCC M 2017556. The initial seed medium was tryptic soy broth (TSB), containing (per liter): tryptone 17 g, soya peptone 3 g, NaCl 5 g, K2HPO4 2.5 g, dextrose 2.5 g, pH 7.3 ± 0.2. The initial production medium contained (per liter) soluble starch 10 g, pharmamedia 5 g, CuSO4 0.05 g, NaI 5 mg, CaCO3 2 g, pH adjusted to 7.0 before autoclaving [44]. The Streptomyces sp. CB03234 mutants identified from ribosome engineering were cultured in the TSB seed medium with 1 μg mL−1 rifamycin (Rif). An aliquot of fresh spore suspension of Streptomyces sp. CB03234 or mutants (about 1 × 107 spores) was inoculated into 50 mL TSB in 250 mL shaking flasks and grown for 36 h at 30 °C and 250 rpm. Then 5 mL of seed culture was transferred to 50 mL production medium in a 250-mL flask and cultured for 6–9 days at 30 °C and 230–250 rpm.
Tiancimycin A was analyzed on a waters ultra performance liquid chromatography (UPLC) system equipped with a PDA detector and an ACQUITY UPLC (Waters), C18 column (2.7 μm, 4.6 mm × 50 mm, Waters). The mobile phase consisted of buffer A (ultrapure H2O containing 0.1% HCOOH and 0.1% CH3CN) and buffer B (chromatographic grade CH3CN containing 0.1% HCOOH) was applied at a flow rate of 0.4 mL min−1. A linear gradient program (60% buffer A and 40% buffer B to 50% buffer A and 50% buffer B for 2 mins, 50% buffer A and 50% buffer B to 0% buffer A and 100% buffer B for 5 mins, followed by 60% buffer A and 40% buffer B for 5 mins) was applied to detect TNM A at 540 nm.
In vitro and in situ resin adsorption of TNM A
The resins HP20, HP2MGL, XAD-16, XAD-2, IRC-50, A30-B were washed prior to use based on the recommendation from the manufacturers. About 1% (w/v) of each type of resins (0.4 g) was added into Streptomyces sp. CB03234 fermentation supernatant (40 mL), respectively, mixing on a rotary shaker for 2 h at room temperature. After the resins were washed by deionized water (30 mL × 3), TNM A was eluted with methanol (MeOH) (30 mL × 3) from HP20, HP2MGL, XAD-16 and XAD-2, or with 20% (w/v) NaCl (10 mL × 3) from IRC-50 and A30-B, which was extracted by ethyl acetate (EtOAc) (30 mL × 3). The organic fractions were then dried in anhydrous Na2SO4, concentrated in vacuum and quantified via UPLC analysis based on the TNM A standard curve (Fig. S1). TNM A in the supernatant before and after resin treatment was extracted by EtOAc, dried in anhydrous Na2SO4, concentrated and similarly quantified. The absorption and deabsorption ratio, and the recovery ratio were then calculated.
The HP2MGL resin was chosen for the subsequent in situ resin addition experiments. Different amounts of HP2MGL resins (0.25, 0.5, 1, 2 and 3%, w/v) were added into 50 mL production medium in 250 mL shaking flasks before autoclaving. The Streptomyces sp. CB03234 strain was then fermented using the resin-containing production medium for 7 days, with production medium without resins as a control. The resins and the supernatant were separated, and TNM A was recovered as above and quantified using UPLC. TNM A in mycelium was obtained by treatment of the mycelium using MeOH (30 mL × 3). The extracts were then dried in anhydrous Na2SO4, concentrated in vacuum, and similarly quantified by UPLC.
Generation of Streptomyces sp. CB03234 mutants by ribosome engineering
The Streptomyces sp. CB03234 wild-type strain was subjected to varying concentrations of Rif to isolate mutants by the ribosome engineering approach alone or in combination with random mutagenesis using diethyl sulfate (DES). The minimum inhibitory concentration (MIC) of Rif against Streptomyces sp. CB03234 was first determined using the agar dilution method [42]. Thus, about 1 × 109 Streptomyces sp. CB03234 spores were treated with 0.46 M DES for 10–40 min at room temperature, resulting in a killing ratio of > 99.9%. Both the non-treated and DES-treated spores (1 × 109) were plated onto 12 GYM plates (per liter: glucose 4 g, yeast extract 4 g, malt extract 10 g, CaCO3 2 g, agar 20 g, pH 7.2), supplemented with 2, 5 or 10 μg mL−1 Rif, respectively, and incubated at 30 °C. Rif-resistant colonies were observed after 6–7 days and confirmed by re-growing them on GYM plates containing 5 μg mL−1 Rif. Chromosomal DNA was extracted and purified from the Streptomyces sp. CB03234 wild-type and Rif-resistant mutant strains following the standard protocol. To amplify the segment of rpoB, PCR experiments were performed by following the manufacturer’s instructions and using Taq polymerase (Takara Ex Taq). An Eppendorf thermal cycler was used, and the PCR conditions were 5 min of incubation at 96 °C, followed by 30 cycles of 96 °C for 1 min, 60 °C for 1 min, and 72 °C for 2 min. The forward and reverse primers were 5′-CCCGAAGCGCTACGACCTCGC-3′ and 5′-GAGGCGAGCGAGCCGATCAGAC-3′. The resulting PCR fragments were cloned into pMD18T (Takara) and subjected to DNA sequencing.
Time course analysis of TNM A production in Streptomyces sp. CB03234 and CB03234-R-16, and the gene expression analysis by quantitative real-time RT-PCR
For the time course analysis of the Streptomyces sp. CB03234 wild-type and the CB03234-R-16 mutant over the 7-day fermentation, the respective cultures (3 × 50 mL) were individually harvested every day. TNM A in the resins was similarly quantified. The pH of the supernatant was measured on a pH meter. The mycelium of each culture was dried and weighed.
The mycelia from about 200 μL culture from the above fermentation at 48, 120 and 168 h were used for total RNA extraction using the total RNA extraction kit (Roche), according to the manufacturer’s instruction. Genomic DNA was removed by RNase-free DNase I (TaKaRa), and the concentration and quality of total RNA were determined using a Biophotometer Nanodrop (Analytik Jena). Reverse transcription was conducted using the cDNA Synthesis Kit (Roche). Quantitative real-time RT-PCR (qRT-PCR) was carried out on the Roche LightCycler 96 fluorescence quantitative PCR system using the Fast Start Essential DNA Green Master (Roche).
The PCR conditions consisted of one cycle of denaturation at 95 °C for 10 min, followed by 40 cycles of 15 s at 95 °C and 60 s at 55 °C. Each transcription assay was normalized to the corresponding transcriptional level of hrdB, and all qRT-PCR reactions were performed in triplicate. The primers used for qRT-PCR were listed in Table S3 in the Supporting Information.
Fermentation medium development
Based on the original production medium, 13 types of carbon sources, including soluble starch (the original carbon source), corn flour, dextrin, d-fructose, d-galactose, d-maltose, d-xylose, glycerol, glucose, α-lactose, mannitol, nonfat milk powder, sucrose, were individually tested at 10 g L−1. Then 14 nitrogen sources including pharmamedia (the original nitrogen source), beef extract, casamino acids, corn steep liquor, corn protein powder, fish peptone, fish meal, malt extract, oat meal, peptone, peanut protein powder, soya peptone, tryptone, and yeast extract, were individually tested at 5 g L−1, the Streptomyces sp. CB03234-R-16 mutant was cultured as before in the production medium without addition of HP2MGL resins. TNM A concentration in the above supernatants was estimated on day 2, 4 and 6 of the fermentation, by the agar diffusion assay against Kocuria rhizophila ATCC 9341.
Four different concentrations of soluble starch (15, 20, 30, 40 g L−1) and pharmamedia (10, 15, 20, 25 g L−1) were tested, respectively. The Streptomyces sp. 3234-R-16 strain was cultured in the production medium with addition of 1.5% (w/v) HP2MGL resins for 9 days, and TNM A was recovered from the resins and quantified by UPLC. Next the combination of soluble starch and pharmamedia, as well as the inorganic salts, including NaI (5, 15, 20, 25, 30, 35, 40 mg L−1), CuSO4 (0.05, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4 g L−1) and CaCO3 (2, 4, 6, 8, 16 g L−1) were investigated.
Fermentation in 15-L fermentors
A 20 μL Streptomyces sp. CB03234-R-16 spore suspension (about 1 × 109 mL−1) was inoculated into 400 mL TSB seed medium with 1 μg mL−1 Rif in two to four 2 L Erlenmeyer flasks and cultured at 30 °C for 36–48 h. The seed culture with varying amounts was transferred to 15 L fermentors with a working volume of 8.0 L using the following production medium: medium A (per liter: soluble starch 40 g, pharmamedia 20 g, CuSO4 0.2 g, NaI 20 mg, CaCO3 8 g and 2 mL anti-foaming agent, pH 7.0); medium B (per liter: soluble starch 30 g, pharmamedia 15 g, CuSO4 0.15 g, NaI 15 mg, CaCO3 6 g, pH 7.0 and 2 mL anti-foaming agent) and medium C (per liter: soluble starch 30 g, pharmamedia 15 g, CuSO4 0.05 g, NaI 5 mg, CaCO3 2 g, pH 7.0 and 2 mL anti-foaming agent). The Streptomyces sp. CB03234-R-16 was fermented at 30 °C, air flow rate of 200 L/h and air pressure 0.05 MPa for 9 days. When the pH of the fermentation culture reached 7.0, 0.2 M HCl was added dropwise to control the pH of the culture around 7.0. For TNM A analysis from the fermentors, the resins from 50 mL fermentation culture were collected, and TNM A from the resins was quantified by UPLC. Medium C was used to obtain the optimal fermentation parameters. To determine the best volume of inoculum, different amounts of seed culture [(5, 10 and 15% (v/v)] was used. To determine the optimal dissolved oxygen (DO) level, 10% inoculum was used, and the agitation speed was correlated with the DO level.
The partial rpoB sequences from Streptomyces sp. CB03234-R-1 to Streptomyces sp. CB03234-R-18 reported in this manuscript were deposited with the GenBank Database under Accession NO MG241318–MG241335.
Results
In situ resin addition increased the titer of TNM A in Streptomyces sp. CB03234
The addition of solid-phase adsorbents during fermentation has increased the production of many valuable microbial natural products, including anticancer agents epothilones, leinamycin, and two enediynes dynemicin A and esperamicin A1, by avoiding certain feedback inhibition or the cytotoxicity of the products [28]. We first screened 4 macroporous resins ranging from different polymer chemistry, surface area, particle and pore sizes, including HP20, HP2MGL, XAD-16, and XAD-2, and two ion exchange resins, IRC-50 and A-30B. The choice of these resins was judicially based on their ability to adsorb TNM A from the fermentation broth of Streptomyces sp. CB03234 and the ease of TNM A to be eluted after the adsorption (Fig. 2a) (Table S2). Among them, the macroporous resin HP2MGL showed the highest TNM A recovery ratio and was thus chosen for the in situ resin addition.
The use of resin addition to improve TNM A production. a The adsorption, deadsorption and recovery ratio of TNM A from various types of resins. b The effect of different amounts of in situ added HP2MGL resin towards TNM A production in Streptomyces sp. CB03234 in shaking flasks. N.D. not detected, N.A. not applicable. There were three independent trials for the data bars
The addition of 0.5–3% (w/v) of HP2MGL resins in Streptomyces sp. CB03234 fermentation medium had some positive effects on TNM A production in 250-mL shaking flasks (Fig. 2b). In comparison to the fermentation without addition of resins, no TNM A was detected in the supernatant and most TNM A was adsorbed to the resins and subsequently eluted. It seemed that the addition of larger amount of resins was instrumental to sequester TNM A from mycelium, since there was only very small amount of TNM A in the mycelium when 1.0% (v/v) or more resins were added. Overall the addition of 1.5% (w/v) of HP2MGL resins led to the increase of TNM A titer to ~ 0.8 mg L−1, which was about threefold improvement over original titer of ~ 0.3 mg L−1 in the absence of resins. The in situ resin adsorption of TNM A also streamlined the isolation procedure, since TNM A could be directly eluted from the resins after fermentation. Therefore, the HP2MGL resins were used in the following experiments.
Ribosome engineering of Streptomyces sp. CB03234 leading to the isolation of the Streptomyces sp. CB03234-R-16 mutant that overproduces TNM A
Ribosome engineering alone, or in combination with random mutagenesis, is a rational method to increase the yield of many secondary metabolites, such as actinorhodin, erythromycin, milbemycin, sinefungin and fredericamycin A [12, 14, 39, 40, 43, 45]. Rif is a potent and selective antibiotic to inhibit DNA-dependent RNA polymerases in bacteria, and has often been used in ribosome engineering [10, 25]. We adopted a strategy to screen for Rif-resistant Streptomyces sp. CB03234 mutants, using spores freshly treated by chemical mutagen diethyl sulfate (DES) or spores untreated, to identify strains with higher TNM A production. First, the MIC of Streptomyces sp. CB03234 towards Rif was determined to be 1 μg mL−1. The spores were then spread onto GYM plates with varying concentrations of Rif, ranging from 2 to 10 μg mL−1. A total of 18 Rif-resistant colonies, named Streptomyces sp. CB03234-R-1 to CB03234-R-18, were isolated and their MICs towards Rif were subsequently determined (Table 1). The MICs for all the isolated mutants towards Rif were at least 5 μg mL−1, five times higher than that for the wild-type. Interestingly, there were several mutants with MICs above 200 μg mL−1. The Rif-resistance is often conferred by mutation in the rpoB gene, which encodes the RNA polymerase β-subunit [26]. We next cloned the rpoB genes of the Streptomyces sp. CB03234 wild-type and the mutants, and subjected them to DNA sequencing. Surprisingly, all mutants except one contain various type of mutation in rpoB, featuring a very high mutation ratio comparing to other reported Rif mutants. Besides the typical mutations on RpoB often identified from Rif-resistant Streptomyces species, such as L422P, D427V, H437Y, R440H and S442L (numbered based on the RpoB from S. coelicolor), there were two new mutations, H437N (Streptomyces sp. CB03234-R-10), and a deletion of a stretch of seven amino acids (FMDQNNP) from RpoB (Streptomyces sp. CB03234-R-5), identified from above mutants (Figs. S2, S3). Note that the H437Y mutation on RpoB was often the most effective mutation in enhancing the antibiotic production in various actinomycetes [25].
We next evaluated TNM A production in mutants bearing different type of mutations, in comparison to the wild-type strain, with the addition of HP2MGL resin in the production medium (Table 1 and Fig. S4). The mutants bearing L422P, D427V, H437N and H437Y mutations on RpoB, showed drastic increase of TNM A production, ranging from 1.5 ± 0.2 to 5.8 ± 1.0 mg L−1, while the others showed no or attenuated TNM A production. Comparing to the wild-type strain with a titer of about 0.8 mg L−1, the highest TNM A producing mutant Streptomyces sp. CB03234-R-16 with L422P mutation showed about 6–7 times improvement, which was selected for the following experiments.
qRT-PCR analysis of Streptomyces sp. CB03234 wild-type and Streptomyces sp. CB03234-R-16 mutant strains revealing drastic increase of gene expression governing TNM A production
A time course analysis of TNM A production in both the Streptomyces sp. CB03234 wild-type and Streptomyces sp. CB03234-R-16 mutant strains was performed (Figs. 3, S5). The effects of RpoB (L422P) mutation on the transcription of relA, which encodes ppGpp synthetase, and selected genes governing TNM A biosynthesis, were assessed at mRNA level using qRT-PCR. The transcription of hrdB, which encodes the principal sigma factor of RNA polymerase, was used as the internal control [5].
The time course analysis of TNM A production in Streptomyces sp. CB03234 wild-type and CB03234-R-16 mutant, and qRT-PCR analysis revealed the overexpression of key genes in Streptomyces sp. CB03234-R-16, comparing to the wild-type. a The biosynthetic gene cluster of TNM A in Streptomyces sp. CB03234 and the genes [5] evaluated in qRT-PCR were highlighted in red. b The production time course of TNM A in Streptomyces sp. CB03234 and Streptomyces sp. CB03234-R-16 in shaking flasks. c–j qRT-PCR analysis of relA and selected genes in TNM A biosynthetic gene cluster. The white bar stands for Streptomyces sp. CB03234 wild-type, and the red bar stands for CB03234-R-16 mutant. hrdB was used as an internal control. tnmL, cytochrome P450 monooxygenase; tnmR7, AraC family transcriptional regulator; tnmR3, unknown protein; tnmE10, type II thioesterase; tnmR1, HxlR family transcriptional regulator; tnmR2, putative regulator; tnmR4, AraC family transcriptional regulator. Each data point represents the mean ± SD of three biological replicates (color figure online)
The expression of relA gene was mainly elevated at the late stages of fermentation in the mutant in comparison to the wild-type strain, as exemplified by a 1.8-fold increase at 120 h and over 100-fold increase at 168 h, with the production of TNM A of 6.6 ± 1.0 or 5.8 ± 1.0 mg L−1 in the mutant, in comparison to 0.9 ± 0.1 or 0.8 ± 0.1 mg L−1 in the wild-type (Fig. 3b, c). This was consistent with the previous observation that certain ribosomal mutations would increase relA transcription and lead to antibiotic overproduction, although the intracellular ppGpp level was not measured in the present study [40]. The transcript amounts of tnmL (cytochrome P450 monooxygenase) and tnmE10 (type II thioesterase) were very abundant, reflecting its direct involvement in TNM A biosynthesis (Fig. 3d, e). For example, the type II thioesterase TnmE10 might play a key role to remove aberrant or stalled biosynthetic intermediates in enediyne polyketide synthases, especially during the beginning of TNM A biosynthesis, since both the wild-type and mutant showed highest tnmE10 expression at 48 h [9, 11, 13]. The constant overexpression of tnmL might reflect the requirement for oxidative modification in furnishing the final natural product, due to the presence of the hydroxylation in either the anthraquinone moiety or the enediyne core in TNM A (Fig. 1). For all the transcriptional factors examined, tnmR4 and tnmR7, both encoding AraC family transcriptional regulator, and tnmR2, encoding a putative regulator, showed about threefold increase of the expression at 168 h in the mutant in comparison to the wild-type strain, while the other examined transcriptional factors showed slightly less variation (Fig. 3f, j). These findings are consistent with previous studies on enediyne biosynthesis and regulation, and the homologues of TnmR4 and TnmR7 have been shown as positive regulators to improve C-1027 production in S. globisporus [6].
Medium optimization leading to overproduction of TNM A in Streptomyces sp. CB03234-R-16
During the 7-day fermentation in the original production medium, the Streptomyces sp. CB03234-R-16 mutant grew slower than the wild-type strain and produced less mycelia. In addition, some degradation of TNM A was also observed with the elevation of the pH in the culture, and the presence of the labile enediyne functional group might affect the stability of TNM A under basic conditions [23, 37] (Figs. 3b, S5). Therefore, we decided to develop an optimal medium for TNM A production in the Streptomyces sp. CB03234-R-16 mutant. Starting from the original medium used in the wild-type, we first evaluated 13 different carbon sources and 14 different nitrogen sources in shaking flasks without addition of the resins. The production of TNM A was monitored at day 2, 4, and 6, respectively, using agar diffusion test with K. rhizophila ATCC 9341 as the indication strain (Fig. S6). The original carbon source and nitrogen source, soluble starch and pharmamedia, were still among the best to produce TNM A in Streptomyces sp. CB03234-R-16.
Then various concentrations of soluble starch, ranging from 10 to 40 g L−1, and pharmamedia, ranging from 10 to 25 g L−1, were individually investigated in the presence of 1.5% (w/v) HP2MGL resins. The increased concentration of soluble starch or pharmamedia in the production medium was beneficial to the growth of Streptomyces sp. CB03234-R-16, but delayed the production of TNM A. However, TNM A titers reached to 9.6 ± 0.2 or 11.1 ± 0.4 mg L−1 when 30 g L−1 of soluble starch or 15 g L−1 pharmamedia was used, compared to 4.4 ± 0.9 mg L−1 in the original medium after 9-day fermentation (Fig. 4a). Finally, the optimal fermentation medium to achieve the highest TNM A titer of 22.5 ± 3.1 mg L−1 in Streptomyces sp. CB03234-R-16 in shaking flasks was the appropriate combination of soluble starch, pharmamedia, and certain inorganic salts, which consisted of 40 g L−1 soluble starch, 20 g L pharmamedia, 8 g L−1 CaCO3, 200 mg L−1 CuSO4·5H2O, 20 mg L−1 NaI and 15 g L−1 HP2MGL resin (Fig. 4b).
Medium optimization of Streptomyces sp. CB03234-R-16 for TNM A overproduction in shaking flasks. a Production of TNM A in different concentrations of soluble starch or pharmamedia. b Production of TNM A in different combinations of soluble starch, pharmamedia and inorganic salts, including NaI, CuSO4 and CaCO3. There were at least three independent trials for the data bars
Production of TNM A from Streptomyces sp. CB03234-R-16 in 15 L fermentors
Production of TNM A from Streptomyces sp. CB03234-R-16 was first scaled up in 15 L fermentors using the optimal production medium based on the shaking flask experiments. But we quickly noticed that TNM A production could only reach to about 10 mg L−1 in 15 L fermentors, mainly due to the high viscosity of the medium and the release of TNM A from some broken HP2MGL resins resulting from agitation (data not shown). Therefore, an alternative medium with less viscosity was selected, consisting of 30 g L−1 starch soluble, 15 g L−1 pharmamedia, 2 g L−1 CaCO3, 50 mg L−1 CuSO4·5H2O, 20 mg L−1 NaI, and 2 mL L−1 anti-foaming agent, along with 3% (w/v) HP2MGL resins to offset the broken resins. We evaluated the effects of inoculation volume of the seed culture (5, 10 and 15%), and DO levels (10–20, 20–60 and 60–80%) towards TNM A production (Fig. S7). We discovered that inoculation of 10% (v/v) of the seed culture and DO level at 20–60% would be optimal for TNM A production. Under the optimal fermentation conditions, the titers of TNM A in Streptomyces sp. CB03234-R-16 reached to 13.0 ± 1.0 mg L−1 (Fig. 5).
The production of TNM A from Streptomyces sp. CB03234-R-16 in 15 L fermentors. There were at least two independent trials for the data bars. The medium consisted of 30 g L−1 starch soluble, 15 g L−1 pharmamedia, 2 g L−1 CaCO3, 50 mg L−1 CuSO4·5H2O, 20 mg L−1 NaI, and 2 mL L−1 anti-foaming agent, along with 2% (v/w) HP2MGL resins
Discussion
Natural products are main sources of anticancer and anti-infective drugs, but one of the main hurdles to the development of natural products as clinical drugs is their limited sources [20, 33]. For microbial drug discovery, many microorganisms can be isolated, preserved, and cultured, thereby offering potential of reliable supply by microbial fermentation. However, the initial titers of most microbial natural products are typically not high enough to warrant their future development. Probably due to the extremely toxicity and instability of the enediyne core, the titers of enediyne natural products are often extremely low, even for the industrial producers, in comparison to many other antibiotics of microbial origin (Fig. 1 and Table S1).
In this study, we have used a combinational approach to rapidly increase the titer of TNM A, a newly discovered enediyne natural product. Ribosome engineering of Streptomyces sp. CB03234 resulted the Streptomyces sp. CB03234-R-16 strain with L422P mutation on RpoB, which afforded a TNM A titer of 5.8 ± 1.0 mg L−1 aided by the in situ added macroporous resins (Table 1 and Fig. S5). Further fermentation optimization for Streptomyces sp. CB03234-R-16 mutant resulted in a TNM A titer of 22.5 ± 3.1 mg L−1 in shaking flasks, and 13.0 ± 1.0 mg L−1 in 15 L fermentors, an increase of ~ 60-fold or ~ 40-fold in comparison to the original titer in the wild-type, respectively (Figs. 4, 5). Optimal concentration of soluble starch, pharmamedia, and inorganic salts in the production medium was critical for higher TNM A production in Streptomyces sp. CB03234-R-16 in both the shaking flasks and 15 L fermentors. The original medium only contained 10 g L−1 soluble starch and 5 g L−1 pharmamedia, and showed highest TNM A production on day 5 (Fig. 3b). In contrast, the optimized nutrient rich medium contained 40 g L−1 soluble starch and 20 g L−1 pharmamedia in shaking flasks, and was thus advantageous for its growth, but delayed the production of TNM A (Fig. 4). However, the higher concentration of 40 g L−1 soluble starch and 20 g L−1 pharmamedia attenuated TNM A production in 15 L fermentors, probably due to the high viscosity in the fermentation medium and the insufficient oxygen supply. Further increase of the agitation speed was undesirable because it would break the macroporous resins in the medium. Therefore, an alternative production medium with less viscosity was finally selected. Our rapid improvement in TNM A titer was largely due to the use of the combinational approach, including ribosome engineering, in situ resin adsorption, medium optimization and fermentation engineering, which should guide the future strain improvement efforts for other enediyne production [44].
The success of ribosome engineering depended critically on the discovery of the so-called “antibiotic overproduction mutations” on RpoB, which have often been associated with increased secondary production [25]. In our study, a total of 17 out of 18 Rif-resistant Streptomyces sp. CB03234 mutants were obtained through ribosome engineering, and there were seven different types of mutations discovered (Table 1 and Fig. S3). The Streptomyces sp. CB03234 mutants baring L422P and H437Y mutations on RpoB showed drastic increase in TNM A production, which have been found to increase the production of antibiotics from several other Streptomyces species, such as actinorhodin in both S. coelicolor and S. lividans, actinomycin in S. parvulus and S. antibioticus, formycin in S. lavendulae, piperidamycin in S. mauvecolor [25]. However, to our knowledge, H437N mutation in RpoB was reported to increase secondary metabolite production in Streptomyces for the first time. In addition to the understanding of the regulation of RNA polymerase by the pleiotropic second messenger ppGpp, a recent structure study has revealed the details of how RelA interacts with the stalled ribosome under amino acid starvation conditions, to initiate the synthesis of (p)ppGpp [4, 29, 38, 46]. Therefore, the observed overexpression of relA is consistent with the marked improved TNM A titer in Streptomyces sp. CB03234-R-16 mutant (Fig. 3). The mutated RNA polymerase in Streptomyces sp. CB03234-R-16 might also have an enhanced affinity towards the promoter regions of the TNM biosynthetic genes, which eventually leads to the TNM A overproduction, in collaborating with the ppGpp effects [12].
The strain Streptomyces sp. CB03234-R-5, with a seven amino acid deletion of FMDQNNP in RpoB, abolished TNM A production, while had a MIC of at least 200 μg mL−1 towards Rif. Shorter deletion of RpoB has been discovered before, but how the mutant survived and how the removal of the seven amino acids in RpoB impacted the function the RNA polymerase remained an interesting question. Along with the availability of the various rpoB mutants from Rif-resistant screening and many other mutations on ribosome discovered from ribosome engineering, rationally designed RpoB should facilitate the future metabolic engineering efforts for secondary metabolites production in microorganisms.
The yield of TNM A could be further improved by metabolic engineering of pathway specific regulators, such as the overexpression of putative transcriptional activators TnmR4 and TnmR7 in Streptomyces sp. CB03234-R-16 mutant, or screening for other antibiotic resistant mutants using the current Rif-resistant mutants. However, our current TNM A titer might already warrant the future pilot-scale production of TNM A, since gram quantity of TNM A would be sufficient to set the stage for preclinical testing of TNM A as the payload for the next generation of anticancer ADCs.
References
Adrio JL, Demain AL (2006) Genetic improvement of processes yielding microbial products. FEMS Microbiol Rev 30:187–214. https://doi.org/10.1111/j.1574-6976.2005.00009.x
Amadori S, Suciu S, Selleslag D, Aversa F, Gaidano G, Musso M, Annino L, Venditti A, Voso MT, Mazzone C, Magro D, De Fabritiis P, Muus P, Alimena G, Mancini M, Hagemeijer A, Paoloni F, Vignetti M, Fazi P, Meert L, Ramadan SM, Willemze R, de Witte T, Baron F (2016) Gemtuzumab Ozogamicin Versus Best Supportive Care in Older Patients with newly diagnosed acute myeloid leukemia unsuitable for Intensive chemotherapy: results of the randomized phase III EORTC-GIMEMA AML-19 trial. J Clin Oncol 34:972–979. https://doi.org/10.1200/JCO.2015.64.0060
Baltz RH (2016) Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other Actinomycetes. J Ind Microbiol Biotechnol 43:343–370. https://doi.org/10.1007/s10295-015-1682-x
Brown A, Fernández IS, Gordiyenko Y, Ramakrishnan V (2016) Ribosome-dependent activation of stringent control. Nature 534:277–280. https://doi.org/10.1038/nature17675
Chakraburtty R, Bibb M (1997) The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J Bacteriol 179:5854–5861. https://doi.org/10.1128/jb.179.18.5854-5861.1997
Chen Y, Yin M, Horsman GP, Shen B (2011) Improvement of the enediyne antitumor antibiotic C-1027 production by manipulating its biosynthetic pathway regulation in Streptomyces globisporus. J Nat Prod 74:420–424. https://doi.org/10.1021/np100825y
Davies J, Wang H, Taylor T, Warabi K, Huang XH, Andersen RJ (2005) Uncialamycin, a new enediyne antibiotic. Org Lett 7:5233–5236. https://doi.org/10.1021/ol052081f
Döhner H, Weisdorf DJ, Bloomfield CD (2015) Acute, myeloid leukemia. N Engl J Med 373:1136–1152. https://doi.org/10.1056/NEJMra1406184
Doi-Katayama Y, Yoon YJ, Choi CY, Yu TW, Floss HG, Hutchinson CR (2000) Thioesterases and the premature termination of polyketide chain elongation in Rif B biosynthesis by Amycolatopsis mediterranei S699. J Antibiot 53:484–495. https://doi.org/10.7164/antibiotics.53.484
Floss HG, Yu TW (2005) Rifamycin-mode of action, resistance, and biosynthesis. Chem Rev 105:621–632. https://doi.org/10.1021/cr030112j
Heathcote ML, Staunton J, Leadlay PF (2001) Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem Biol 8:207–220. https://doi.org/10.1016/S1074-5521(01)00002-3
Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K (2009) Antibacterial discovery in Actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol 27:462–464. https://doi.org/10.1038/nbt.1538
Kotowska M, Pawlik K (2014) Roles of type II thioesterases and their application for secondary metabolite yield improvement. Appl Microbiol Biotechnol 98:7735–7746. https://doi.org/10.1007/s00253-014-5952-8
Lai C, Xu J, Tozawa Y, Okamoto-Hosoya Y, Yao X, Ochi K (2002) Genetic and physiological characterization of rpoB mutations that activate antibiotic production in Streptomyces lividans. Microbiology 148:3365–3373. https://doi.org/10.1099/00221287-148-11-3365
Lam KS, Gustavson DR, Veitch JA, Forenza S (1993) The effect of cerulenin on the production of esperamicin A1 by Actinomadura verrucosospora. J Ind Microbiol 12:99–102. https://doi.org/10.1007/BF01569908
Lam KS, Veitch JA, Lowe SE, Forenza S (1995) Effect of neutral resins on the production of dynemicins by Micromonospora chersina. J Ind Microbiol 15:453–456. https://doi.org/10.1007/BF01569975
Lam KS, Titus JA, Dabrah TT, Kimball DL, Veitch JM, Gustavson DR, Compton BJ, Matson JA, Forenza S, Ross J, Miller D, Roach J, Beutler J (1992) Improved processes for the production and isolation of dynemicin A and large-scale fermentation in a 10,000-liter fermentor. J Ind Microbiol 11:7–12. https://doi.org/10.1007/BF01583725
Lamb YN (2017) Inotuzumab ozogamicin: first global approval. Drugs. https://doi.org/10.1007/s40265-017-0802-5
Maeda H, Konno T (1997) Metamorphosis of neocarzinostatin to SMANCS: chemistry, biology, pharmacology, and clinical effect of the first prototype anticancer polymer therapeutic. In: Maeda H, Edo K, Ishida N (eds) Neocarzinostatin. Springer, Tokyo. https://doi.org/10.1007/978-4-431-66914-2_12
Newman DJ, Cragg GM (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79:629–661. https://doi.org/10.1021/acs.jnatprod.5b01055
Nicolaou KC, Wang Y, Lu M, Mandal D, Pattanayak MR, Yu R, Shah AA, Chen JS, Zhang H, Crawford JJ, Pasunoori L, Poudel YB, Chowdari NS, Pan C, Nazeer A, Gangwar S, Vite G, Pitsinos EN (2016) Streamlined total synthesis of uncialamycin and its application to the synthesis of designed analogues for biological investigations. J Am Chem Soc 138:8235–8246. https://doi.org/10.1021/jacs.6b04339
Nicolaou KC, Lu Z, Li R, Woods JR, Sohn TI (2015) Total synthesis of shishijimicin A. J Am Chem Soc 137:8716–8719. https://doi.org/10.1021/jacs.5b05575
Nicolaou KC, Dai WM (1991) Chemistry and biology of the enediyne anticancer antibiotics. Angew Chem Int Ed Engl 30:1387–1530. https://doi.org/10.1002/anie.199113873
Nielsen J, Keasling JD (2016) Engineering cellular metabolism. Cell 164:1185–1197. https://doi.org/10.1016/j.cell.2016.02.004
Ochi K (2016) Insights into microbial cryptic gene activation and strain improvement: principle, application and technical aspects. J Antibiot 70:25–40. https://doi.org/10.1038/ja.2016.82
Ochi K (2007) From microbial differentiation to ribosome engineering. Biosci Biotechnol Biochem 71:1373–1386. https://doi.org/10.1271/bbb.70007
Olano C, Lombó F, Méndez C, Salas JA (2008) Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng 10:281–292. https://doi.org/10.1016/j.ymben.2008.07.001
Phillips T, Chase M, Wagner S, Renzi C, Powell M, DeAngelo J, Michels P (2013) Use of in situ solid-phase adsorption in microbial natural product fermentation development. J Ind Microbiol Biotechnol 40:411–425. https://doi.org/10.1007/s10295-013-1247-9
Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL (2013) The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell 50:420–429. https://doi.org/10.1016/j.molcel.2013.03.021
Santos CN, Stephanopoulos G (2008) Combinatorial engineering of microbes for optimizing cellular phenotype. Curr Opin Chem Biol 12:168–176. https://doi.org/10.1016/j.cbpa.2008.01.017
Schlenk RF, Müller-Tidow C, Benner A, Kieser M (2017) Relapsed/refractory acute myeloid leukemia: any progress? Curr Opin Oncol 29:467–473. https://doi.org/10.1097/CCO.0000000000000404
Shao RG, Zhen YS (2008) Enediyne anticancer antibiotic lidamycin: chemistry, biology and pharmacology. Anticancer Agents Med Chem 8:123–131. https://doi.org/10.2174/187152008783497055
Shen B (2015) A new golden age of natural products drug discovery. Cell 163:1297–1300. https://doi.org/10.1016/j.cell.2015.11.031
Shen B, Hindra Yan X, Huang T, Ge H, Yang D, Teng Q, Rudolf JD, Lohman JR (2015) Enediynes: exploration of microbial genomics to discover new anticancer drug leads. Bioorg Med Chem Lett 25:9–15. https://doi.org/10.1016/j.bmcl.2014.11.019
Shen B, Liu W, Nonaka K (2003) Enediyne natural products: biosynthesis and prospect towards engineering novel antitumor agents. Curr Med Chem 10:2317–2325. https://doi.org/10.2174/0929867033456701
Sievers EL, Senter PD (2013) Antibody-drug conjugates in cancer therapy. Annu Rev Med 64:15–29. https://doi.org/10.1146/annurev-med-050311-201823
Smith AL, Nicolaou KC (1996) The enediyne antibiotics. J Med Chem 39:2103–2117. https://doi.org/10.1021/jm9600398
Talà A, Wang G, Zemanova M, Okamoto S, Ochi K, Alifano P (2009) Activation of dormant bacterial genes by Nonomuraea sp. strain ATCC 39727 mutant-type RNA polymerase. J Bacteriol 191:805–814. https://doi.org/10.1128/JB.01311-08
Tanaka Y, Kasahara K, Hirose Y, Murakami K, Kugimiya R, Ochi K (2013) Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance (rpoB) mutations in Actinomycetes. J Bacteriol 195:2959–2970. https://doi.org/10.1128/JB.00147-13
Wang G, Hosaka T, Ochi K (2008) Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl Environ Microbiol 74:2834–2840. https://doi.org/10.1128/AEM.02800-07
Weber T, Charusanti P, Musiol-Kroll EM, Jiang X, Tong Y, Kim HU, Lee SY (2015) Metabolic engineering of antibiotic factories: new tools for antibiotic production in Actinomycetes. Trends Biotechnol 33:15–26. https://doi.org/10.1016/j.tibtech.2014.10.009
Wiegand I, Hilpert K, Hancock REW (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. https://doi.org/10.1038/nprot.2007.521
Xu J, Tozawa Y, Lai C, Hayashi H, Ochi K (2002) A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3(2). Mol Genet Genomics 268:179–189. https://doi.org/10.1007/s00438-002-0730-1
Yan X, Ge H, Huang T, Hindra Yang D, Teng Q, Crnovčić I, Li X, Rudolf JD, Lohman JR, Gansemans Y, Zhu X, Huang Y, Zhao LX, Jiang Y, Van Nieuwerburgh F, Rader C, Duan Y, Shen B (2016) Strain prioritization and genome mining for enediyne natural products. MBio 7:e02104–e02116. https://doi.org/10.1128/mBio.0210416
Zhang Y, Huang H, Xu S, Wang B, Ju J, Tan H, Li W (2015) Activation and enhancement of fredericamycin A production in deepsea-derived Streptomyces somaliensis SCSIO ZH66 by using ribosome engineering and response surface methodology. Microb Cell Fact 14:64. https://doi.org/10.1186/s12934-015-0244-2
Zuo Y, Wang Y, Steitz TA (2013) The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell 50:430–436. https://doi.org/10.1016/j.molcel.2013.03.020
Acknowledgements
This work was supported in parts by NSFC Grants 81473124 (to Y.H.), 81530092 (to B.S.), the Chinese Ministry of Education 111 Project B0803420 (to Y.D.), NIH Grants GM115575 and CA204484 (to B.S.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This article does not contain any studies with human participants performed by any of the authors.
Conflict of interest
All authors declare that he/she has no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, L., Pan, J., Wang, Z. et al. Ribosome engineering and fermentation optimization leads to overproduction of tiancimycin A, a new enediyne natural product from Streptomyces sp. CB03234. J Ind Microbiol Biotechnol 45, 141–151 (2018). https://doi.org/10.1007/s10295-018-2014-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-2014-8