Abstract
In a cell-surface display (CSD) system, successful display of a protein or peptide is highly dependent on the anchoring motif and the position of the display in that anchoring motif. In this study, a recombinant bacterial CSD system for manganese (Mn) and cobalt (Co) recovery was developed by employing OmpC as an anchoring motif on three different external loops. A portion of Cap43 protein (TRSRSHTSEG)3 was employed as a manganese and cobalt binding peptide (MCBP), which was fused with OmpC at three different external loops. The fusions were made at the loop 2 [fusion protein-2 (FP2)], loop 6 (FP6), and loop 8 (FP8) of OmpC, respectively. The efficacy of the three recombinant strains in the recovery of Mn and Co was evaluated by varying the concentration of the respective metal. Molecular modeling studies showed that the short trimeric repeats of peptide probably form a secondary structure with OmpC, thereby giving rise to a difference in metal recovery among the three recombinant strains. Among the three recombinant strains, FP6 showed increased metal recovery with both Mn and Co, at 1235.14 (1 mM) and 379.68 (0.2 mM) µmol/g dry cell weight (DCW), respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metals are solid substances obtained from the earth. The term metal is used for any substance that is hard, possessing silvery luster and is a good conductor of heat and electricity. Due to their natural properties of rigidity, ductility, and tensile strength, metals are used in various fields. Metals are strong, durable, and highly resistant to natural wear and tear and so have been used by humans since ancient times. The accelerated rate of industrialization increased the demand for metals [22]. Currently, almost any object produced by technology uses metals in some form. The sources of metals are exhaustible natural resources, which are quantitatively used at a rapid rate. On the other hand, metals can be recovered from waste, which is thus considered a secondary raw material for metals. Developing an efficient method to recover metals from waste is essential to protect the environment and to provide a constant supply of metals for humankind. The recycling process does not affect the chemical or physical properties of non-ferrous metals; therefore, these metals can be recycled an infinite number of times [22].
Metal recovery using micro-organisms is gaining greater attention. In recent years, a new discipline of mineral science called bio-hydrometallurgy or microbial mining (mining with microbes) has grown rapidly. Bio-hydrometallurgy deals with the application of biotechnology to the mining industry [36]. Mining with microbes is both economically and environmentally friendly. Micro-organisms utilized for metal recovery typically involve either a bio-leaching or bio-sorption process [32]. Bio-leaching is the process of extraction or solubilization of minerals from ores by micro-organisms [21], whereas bio-sorption deals with cell-surface adsorption of metals by micro-organisms. Bio-sorption can be employed to recover valuable and toxic metals from industrial effluent; however, the sorption is non-specific. Metals can be selectively adsorbed by micro-organisms by employing cell-surface display (CSD) of peptides.
CSD enables the display of proteins, peptides, and enzymes on the cell surface and is widely employed in various fields including microbiology [20, 30], biotechnology [6], and vaccinology [27, 35]. CSD involves the display of a gene for a specific peptide, with the outer membrane proteins of a bacteria acting as an anchoring motif for the displayed protein. Many different anchoring proteins have been developed for surface display, and each carrier results in physiological effects on the host cells [23]. The choice of carrier protein or anchoring motif is highly important because an incorrect motif can destabilize the cell envelope integrity and cause cellular effects. In a bacterial cell-surface display system, various anchoring motifs such as outer membrane proteins, auto transporters, and S-layer proteins have been used [18].
The most commonly used anchoring motifs include OmpA [5], OmpF [26], OmpC [41], LamB [7], Lpp-OmpA [15], PhoE [2], Yiat [18], OmpX [42], ice nucleation protein (INP) [17], and a-agglutinin [19]. The fusion of these outer membrane proteins with a protein of interest enables the display of peptides on the outer membrane of the bacteria. The display of proteins or peptides on the surface of a microorganism by recombinant DNA technology has become an increasingly promising technology for various applications and has been widely used in bioremediation [28, 29, 37, 39]. Although OmpC has been extensively studied for surface display in various applications, each anchoring motif has been found to have different capacities of protein display when exposed to various anchoring motifs with respect to size and characteristics [18]. The location of the peptide in the anchoring protein is important, since it influences the immobilization, stability, specific activity, and post-translational modification of the fusion proteins [23].
Mn (Manganese) is the fourth most used metal followed by iron, aluminium, and copper. Approximately 95% of the globally produced Mn is used in the steel industry for the desulfurization and strengthening of steel, and the remaining 5% is used by the battery and chemical industries [9]. Due to anthropogenic activities, exposure of Mn to surface and ground water is relatively high [16]. Mn-containing material, such as waste batteries and spent electrodes, are some of the sources of Mn that leach into effluent with other ores [10]. Exposure to huge amounts of mine effluent has resulted from widespread mining and untreated anthropogenic and industrial activity [9]. Mn at a concentration greater than 0.05 milligrams per liter in tap water is considered unsafe by the US Environmental Protection Agency (US EPA, 2004) [16]. Mn is recognized as a neurotoxin at high-level exposure, and chronic exposure to mining, welding, and manufacturing anthropogenic effluents of this metal contaminant lead to deleterious health effects including Parkinson’s disease, psychiatric symptoms, neurodegenerative diseases, and other disturbances in the motor system [9].
Co (Cobalt) is an essential element in human health as it is a part of vitamin B12; however, a higher concentration of Co can cause low blood pressure, paralysis, diarrhea, and bone defects [1]. Co is considered to be a rare element that is common in various industrial effluents such as metallurgical, paint, mining, and electronic industrial waste [11]. Co is notable as an activated radioisotope and is particularly harmful due to its long half-life (5.27 years) [12]. Several strategies used for the removal of metals from aqueous solution, including chemical precipitation, ion exchange, and coagulation/flocculation, are ineffective, especially for a lower concentration of pollutant [11, 28]. Thus, there is a need for the development of novel cleanup strategies [28].
In this study, an adsorption system for Mn and Co was developed using three different loops of OmpC as an anchoring motif. OmpC was considered for its high copy level with 2 × 105 molecules per cell, which makes it an efficient candidate for cell-surface display [24]. Among the several outer membrane proteins (OMPs), OmpC is a trimeric porin protein found in the outer membrane of E. coli, and it is structurally important for outer membrane stabilization. Three molecules of OmpC form a pore structure on the E. coli cell membrane, which allows small molecules to pass through [8]. An OmpC molecule is comprised of 16 transmembranes, antiparallel β-strands that form a β-barrel structure surrounding a large channel, which is connected to seven internal loops and eight external loops [41]. The peptide (TRSRSHTSEG)3 is a portion of the Cap43 protein [31] that is employed as a Mn and Co binding peptide (MCBP). This is a multi-histidinic peptide fragment from Cap43 which was believed to bind Mn and Co. The most stricking feature of the protein lies in its C-terminal moiety, where the 10 amino acid TRSRSHTSEG are repeated thrice consecutively. So to enhance the binding nature of the manganese and cobalt binding peptide, the peptides are displayed on the cell-surface display. In that strategies, this trimeric repeats of the peptide TRSRSHTSEG were displayed at loop2 (FP2), loop6 (FP6), and loop8 (FP8) of OmpC. All three CSD systems were evaluated for protein expression and efficiency toward Mn and Co adsorption.
Materials and methods
Bacterial strains and media
The bacterial strains used in this study are listed in Table S1. The strains were cultivated in LB medium (10 g/L bacto-tryptone, 5 g/L bacto-yeast extract, and 5 g/L NaCl) supplemented with antibiotic (100 mg/L ampicillin) at 37 °C with vigorous shaking at 250 rpm.
Design of the engineered strains
An MCBP composed of 30aa (TRSRSHTSEG)3 was fused at the C-terminus of truncated OmpC. Fusion was carried out with three external loops of OmpC: at loop2 189th bp (FP2), loop6 726th bp (FP6), and loop8 993rd bp (FP8). The respective genes were amplified with peptide sequence-incorporated oligonucleotides (Table S2). Polymerase chain reaction (PCR) was performed with an MJ Mini Personal Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) using the Expand High Fidelity PCR system (Roche Molecular Biochemicals, Mannheim, Germany). The three PCR products were cloned into the pBAD30 plasmid using SacI and HindIII restriction enzymes to construct pBADOFP2, pBADOFP6, and pBADOFP8, respectively (Fig. 1). The expression of OmpC along with the peptide was under the control of the PBAD promoter. These plasmids were transformed into a chemically competent XL1-Blue (XB) E. coli strain for further studies.
Expression study by SDS-PAGE
The three recombinant E. coli strains were cultured overnight in LB medium at 37 °C, and the cultures were then diluted 100-fold in the same medium. When the optical density at 600 nm OD600) reached 0.5, arabinose was added to the culture medium at various concentrations from 0 to 0.5% and incubated for 6 h. The harvested recombinant strain was sonicated (30 s ON, 30 s OFF, 12 cycles), and the cells were centrifuged at 8000 rpm to remove cell debris. The outer membrane fractions were isolated from the pellet by adding 10 mM Tris–HCl (pH 7.5), and the suspended cells were kept in 4 °C overnight to lift the membrane fractions into solution, which was analyzed by 12% (w/v) sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Fractioned protein samples were stained with Coomassie brilliant blue R-250 (Bio-Rad Laboratories, Hercules, CA, USA).
Mn and Co adsorption and recovery analysis
The three recombinant strains incorporating pBADOFP2, pBADOFP6, and pBADOFP8 were grown overnight in LB medium supplemented with 100 mg/L ampicillin at 37 °C. The overnight cultures were diluted 100-fold in fresh LB medium and incubated until the OD600 reached 0.5. The cells were induced with 0.5% of arabinose, and the strains were then cultured for 6 h at 30 °C. The strains were washed twice with 0.85% (w/v) NaCl, and the cells were incubated with various concentrations of MnCl2 from 0.1 to 1 mM and separately with CoCl2 at concentrations of 0.01– 0.3 mM. To recover the cell-surface-adsorbed Mn and Co, the strains were washed twice with 0.85% (w/v) NaCl, and the cells were incubated with 5 mM EDTA in ice for 30 min.
The selectivity of the peptide was analyzed with artificially polluted wastewater. The concentration of each metal [Manganese (Mn), Cobalt (Co), Copper (Cu), Lead (Pb), and Lithium (Li)] present in the artificially polluted wastewater was 0.1 mM. The same procedure was followed for the recovery of metals used in LB medium.
The adsorption of the best recombinant strain was analyzed by a JEOL JSM-6500F field emission scanning electron microscope to visualize the adsorbed Mn and Co, with E. coli XB strain as a control. To confirm the elemental composition of the samples after adsorption, energy dispersive spectroscopy (EDS) was used. To prepare for SEM/EDS analysis, the samples after adsorption were lyophilized on an EYELA FDU-2200 freeze dryer and were subsequently analyzed with a JEOL JSM-6500F field emission scanning electron microscope equipped with EDS.
Molecular modeling studies
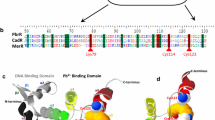
To investigate the cause of the difference in metal binding affinity of MCBP fused at three different loops of OmpC, three-dimensional structures of the three FPs were constructed using the comparative modeling tool Modeler [13]. Further, the full-length sequences of the FPs were aligned with the structural template using an online tool to analyze the secondary structure. Since the three FPs contain partial loops of OmpC protein and MCBP, a fusion model structure was constructed for structural investigation. To develop three-dimensional structures, a structural template search for the full-length sequence FPs and the individual sequences of OmpC and MCBP was performed against the PDB database using the BlastP tool [3]. In addition, multiple sequence alignment was performed with the online tool espript3 [4] using the secondary structure of OmpC as a template (PDB ID-2J1N) [34].
Results and discussion
The surface display can be used for various biotechnological applications such as live vaccine development, screening display peptide libraries, bio-sorbent, whole-cell biocatalyst for remediation of pollutants, and for biofuel production [18]. Until now, many proteins ranging from small peptides to large, complex proteins have been displayed on the surface of various hosts including bacteria, yeast, cyanobacteria, and others [14, 25, 33, 35, 42]. Escherichia coli is the most recurrently used bacterial host for the surface display of proteins and peptides due to its high yield of recombinant proteins and flexibility to genetic manipulation [38]. To attain a proper display of proteins on the cell surface, the system should contain both an inner membrane and an outer membrane, separated by periplasm. The gene of interest to be displayed on the cell surface should be fused to an anchoring protein that enables export across the cell envelope and anchors passengers to the host cell surface. These surface-displayed peptides or proteins are efficiently exposed to the externally added substrate, which enabled the use of cell-surface display as an efficient system for various applications [40]. The most frequently used systems for the expression of proteins or peptides on the cell surface are OMPs. OMPs form a distinct integral membrane-like protein with the common structural motif of a β-barrel, composed of a different number of transmembrane β-strands linked to the periplasmic side and outside with a long surface reachable loop. Many different OMPs have been used for the development of surface display systems including OprF, PhoE, OmpS, OmpF and OmpC [39].
Construction of a Mn and Co binding bacterial surface display system
For the development of an efficient display system, the location in the carrier protein is very important for the effective display of proteins or peptides [23]. OmpC, an outer membrane protein of E. coli, contains eight external loops that can be used for fusion of peptides or proteins [41]. In this work, a C-terminal fusion of MCBP (TRSRSHTSEG)3 at 3 different external loops of OmpC, loop 2, loop 6, and loop 8 (Fig. 1), was constructed. The construction was made possible by truncating ompC at the respective sites (189, 726, 993 bp), followed by fusion of MCBP at the C-terminus of the truncated loops using the respective oligonucleotide (Table S2). The genes were cloned into the pBAD30 vector using SacI and HindIII sites to prepare pBADOFP2, pBADOFP6, and pBADOFP8 (Table S1). The expression of all three recombinant strains was regulated by the PBAD promoter, and protein expression was induced by the addition of arabinose (Fig. 2).
Optimization of surface display conditions
To investigate efficient peptide display conditions, strains containing pBADOFP2, pBADOFP6, and pBADOFP8 were induced with increasing concentrations of arabinose from 0 to 0.5%. The protein expressions of the three FPs were analyzed by SDS-PAGE analysis. The sizes of the FPs were 10 kDa (FP2), 29.7 kDa (FP6), and 39.7 kDa (FP8), respectively (Fig. 3). Since the expression of OmpC is highly influenced by temperature, the expression of the three recombinant strains was studied at two different temperatures (25 and 30 °C) [28]. The expression at 25 °C was very low compared to that at 30 °C for all three strains (results not shown). Based on the above results, optimum conditions for expression of FPs were 30 °C with 0.5% of arabinose for 6 h.
Mn and Co-adsorption studies
The performance of the three recombinant strains was evaluated by incubating the MCBP displayed cells to varying concentrations of MnCl2 and CoCl2. The plasmid not containing an MCBP was used as a control (Table S2). For strains incorporating plasmids pBADOFP2, pBADOFP6, and pBADOFP8, maximum adsorptions of Mn were 586.32, 1235.14, and 1125.24 µmol/g DCW, respectively, from an initial concentration of 1 mM (Fig. 4a–c). The adsorption of Mn increased as its concentration in the medium increased from 0.1 to 1 mM; however, above 3 mM, the adsorption reached a plateau and did not increase further. Based on the above result, Loop 6 showed the highest adsorption toward Mn, followed by loop 2 and loop 8. However, in the three control strains carrying only loop 2, loop 6, or loop 8 of OmpC, the maximum adsorptions were 53, 61, and 78 µmol/g DCW, respectively.
Next, the ability of these recombinant strains to adsorb Co was evaluated by culturing the strains with CoCl2. The same negative and positive control cells used in Mn adsorption studies were used as controls. The recombinant strains harboring pBADOFP2, pBADOFP6, or pBADOFP8 after peptide display exhibited a higher absorption of Co around 236.93, 379.68, and 369.65 µmol/g DCW, respectively, from an initial concentration of 0.2 mM (Fig. 5a–c). The maximum absorptions in the control strains were 43.5, 59.54, and 76.14 µmol/g DCW of Co, respectively. In the case of Co adsorption, FP6 showed higher adsorption ability similar to the results with Mn. Adsorption was relatively low with recombinant cells including plasmids without metal binding peptide compared to the peptide displayed cells (Fig. 5a–c).
The ability of the three recombinant strains to participate in environmental applications and the selectivity of the peptides were further tested in artificially polluted wastewater containing 0.1 mM of the respective metals. All three recombinant strains showed higher selectivity toward Mn, followed by Co. The selectivity of the MCBP toward various metals was in the order Mn > Co > Cu > Pb > Li for all three recombinant strains (Fig. 6a–c). The pBADOFP6 plasmid-incorporated strain had higher Mn and Co adsorptions of approximately 326.325 and 220.153 µmol/g DCW, respectively (Fig. 6b). This result suggests that this 30 aa trimeric repeat peptide (TRSRSHTSEG) could be used for the efficient and selective adsorption of Mn and Co from the environment.
SEM–EDS analysis for pBADOFP6
To provide further evidence of the adsorption of Mn and Co by the recombinant strains, the recombinant strain containing the pBADOFP6 plasmid was subjected to FE-SEM and EDS analyses after adsorption for visual observation of adsorbed metals and composition determination, respectively. The cells after adsorption were washed and visualized using FE-SEM, and the same cells were lyophilized for EDS analysis. Metal adsorption of the pBADOFP6 plasmid-containing strain before and after adsorption is clearly shown in Fig. 7. EDS analysis also clearly showed the presence of Mn and Co in the respective strains after adsorption (Fig. 7e, f).
FE-SEM images of recombinant strains containing MCBP before and after Mn and Co adsorption. a, b The recombinant cells harboring pBADOFP6 before adsorption. c The recombinant strain harboring pBADOFP6 after adsorption with 0.2 mM CoCl2. d The recombinant strain harboring pBADOFP6 after adsorption with 1 mM MnCl2. The arrows indicate adsorption of metals on the cell surface. e EDS analysis of the pBADOFP6 recombinant strain after Co adsorption. f EDS analysis of the pBADOFP6 recombinant strain after Mn adsorption
Molecular modeling studies
The fused MCBP behaves as part of the OmpC loop; hence, the entire peptide is not available for metal binding. In order to validate the difference in adsorption among the three FPs, molecular modeling studies were performed. The metal binding property differs based on efficient peptide display. Preliminary analysis was performed by multiple sequence alignment, as shown in Fig. S1. The FPs were aligned with the full-length OmpC. Based on the alignment results with FP2, the two repeats of MCBP aligned with β sheets 4 and 5, where only a single repeat was freely available as a turn or coil. In the case of FP6, only one repeat was freely available, and the other two were aligned in the middle of β sheets 12 and 13 of OmpC. For FP8, the first and last repeats were aligned with the beginning of B17-alpha helix-B18, and the middle repeat was available for metal binding. To validate the alignment results, the modeled structures of all three FPs were constructed, as shown in Fig. 8, without ignoring the ends using OmpC (2JIN) as a structural template. The modeling studies revealed the difference in peptide display efficiency among the three FPs. In FP2, the first two repeats of MCBP were involved in β sheet formation, and the last only one repeat was involved in metal binding (Fig. 8a). Furthermore, with FP6, two repeats of MCBP were available for metal binding, and only one repeat formed a helix. Though the sequence alignment results showed that the two repeats of FP6 MCBP form a β sheet, the probability that FP6 will turn and to form the same substructure is low (Fig. 8b). In the case of FP8, one and one-half of the MCBP is involved in a helix loop, is responsible for the metal binding property (Fig. 8c). These results clearly indicate that the difference in metal binding activity of the FPs is due to the availability of free loops from MCBP. In addition, the freely available trimeric repeat of MCBP forms a loop that binds metal ions.
Conclusions
The development of a cell-surface display system for whole-cell biocatalysis is highly beneficial for environmental applications. The construction of a cell-surface display system for a specific protein or peptide with a particular anchoring motif has to be examined by both molecular modeling and wet-lab analysis for efficient display of the protein for maximum activity. In this investigation, we developed an efficient system for adsorption of Mn and Co by evaluating various anchoring sites on OmpC. Mn and Co are important industrial minerals. Mn is a metal with important uses in industrial metal alloys, particularly the stainless steel sector. Domestic consumption of Mn is estimated to be 500,000 metric tons/year. The significance of Mn has increased with modern development in steel-making technology. Mn and Co are also employed as electrodes in batteries. It was reported in 2016 that the price of Co would likely rise around 45% by 2020. Thus, it is prudent to design alternative systems to recover these metals. The system developed in this work will be highly efficient in the recovery of Mn and Co from polluted environmental sites.
References
Abbas M, Kaddour S, Trari M (2014) Kinetic and equilibrium studies of cobalt adsorption on apricot stone activated carbon. J Ind Eng Chem 20:745–751
Agterberg M, Adriaanse H, van Bruggen A, Karperien M, Tommassen J (1990) Outer-membrane PhoE protein of Escherichia coli K-12 as an exposure vector: possibilities and limitations. Gene 88:37–45
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Baslé A, Rummel G, Storici P, Rosenbusch JP, Schirmer T (2006) Crystal structure of Osmoporin OmpC from E. coli at 2.0 Å. J Mol Biol 362:933–942
Bessette PH, Rice JJ, Daugherty PS (2004) Rapid isolation of high-affinity protein binding peptides using bacterial display. Protein Eng Des Sel 17:731–739
Binder U, Matschiner G, Theobald I, Skerra A (2010) High-throughput sorting of an anticalin library via EspP-mediated functional display on the Escherichia coli Cell Surface. J Mol Biol 400:783–802
Brown S (1992) Engineered iron oxide-adhesion mutants of the Escherichia coli phage lambda receptor. Proc Natl Acad Sci 89:8651–8655
Cruz N, Le Borgne S, Hernández-Chávez G, Gosset G, Valle F, Bolivar F (2000) Engineering the Escherichia coli outer membrane protein OmpC for metal bioadsorption. Biotechnol Lett 22:623–629
Das AP, Ghosh S, Mohanty S, Sukla LB (2015) Advances in manganese pollution and its bioremediation. In: Sukla LB, Pradhan N, Panda S, Mishra BK (eds) Environmental microbial biotechnology. Springer International Publishing, Switzerland
Das AP, Sukla LB, Pradhan N, Nayak S (2011) Manganese biomining: a review. Bioresour Technol 102:7381–7387
Dotto GL, Cunha JM, Calgaro CO, Tanabe EH, Bertuol DA (2015) Surface modification of chitin using ultrasound-assisted and supercritical CO2 technologies for cobalt adsorption. J Hazard Mater 295:29–36
Duprey A, Chansavang V, Frémion F, Gonthier C, Louis Y, Lejeune P, Springer F, Desjardin V, Rodrigue A, Dorel C (2014) “NiCo Buster”: engineering E. coli for fast and efficient capture of cobalt and nickel. J Biol Eng 8:1–11
Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen M-Y, Pieper U, Sali A (2001) Comparative protein structure modeling using MODELLER. In: Coligan JE (ed) Current protocols in protein science. Wiley
Ferri S, Nakamura M, Ito A, Nakajima M, Abe K, Kojima K, Sode K (2015) Efficient surface-display of autotransporter proteins in cyanobacteria. Algal Res 12:337–340
Georgiou G, Stathopoulos C, Daugherty PS, Nayak AR, Iverson BL, Iii RC (1997) Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat Biotech 15:29–34
Gerke TL, Little BJ, Barry Maynard J (2016) Manganese deposition in drinking water distribution systems. Sci Total Environ 541:184–193
Gurian-Sherman D, Lindow SE (1993) Bacterial ice nucleation: significance and molecular basis. FASEB J 7:1338–1343
Han M-J, Lee SH (2015) An efficient bacterial surface display system based on a novel outer membrane anchoring element from the Escherichia coli protein YiaT. FEMS Microbiol Lett 362:1–7
Jahns AC, Rehm BHA (2012) Relevant uses of surface proteins—display on self-organized biological structures. Microbial Biotechnol 5:188–202
Jin Z, Han S-Y, Zhang L, Zheng S-P, Wang Y, Lin Y (2013) Combined utilization of lipase-displaying Pichia pastoris whole-cell biocatalysts to improve biodiesel production in co-solvent media. Bioresour Technol 130:102–109
Krebs W, Brombacher C, Bosshard PP, Bachofen R, Brandl H (1997) Microbial recovery of metals from solids. FEMS Microbiol Rev 20:605–617
L canda THaEA (2017) Effect of EPR coefficient policy on the production decision in precious metal accessory recycling. Int J Prod Res 55:1129
Lee SY, Choi JH, Xu Z (2003) Microbial cell-surface display. Trends Biotechnol 21:45–52
Li P-S, Tao H-C (2015) Cell surface engineering of microorganisms towards adsorption of heavy metals. Crit Rev Microbiol 41:140–149
Liu Y, Zhang R, Lian Z, Wang S, Wright AT (2014) Yeast cell surface display for lipase whole cell catalyst and its applications. J Mol Catal B Enzym 106:17–25
Madhuranayaki TK (2011) Protein engineering of yellow fluorescent protein insertion in outer membrane protein OmpF from Salmonella typhi. ISABB J Biotechnol Bioinform 1(1):10–14
Majander K, Korhonen TK, Westerlund-Wikström B (2005) Simultaneous display of multiple foreign peptides in the FliD capping and FliC filament proteins of the Escherichia coli Flagellum. Appl environ Microbiol 71:4263–4268
Maruthamuthu M, Nadarajan S, Ganesh I, Ravikumar S, Yun H, I-k Yoo, Hong S (2015) Construction of a high efficiency copper adsorption bacterial system via peptide display and its application on copper dye polluted wastewater. Bioprocess Biosyst Eng 38:2077–2084
Maruthamuthu M, Ganesh I, Ravikumar S, Hong S (2015) Evaluation of zraP gene expression characteristics and construction of a lead (Pb) sensing and removal system in a recombinant Escherichia coli. Biotechnol Lett 37:659–664
Matano Y, Hasunuma T, Kondo A (2013) Cell recycle batch fermentation of high-solid lignocellulose using a recombinant cellulase-displaying yeast strain for high yield ethanol production in consolidated bioprocessing. Bioresour Technol 135:403–409
Peana M, Medici S, Nurchi VM, Crisponi G, Lachowicz JI, Zoroddu MA (2013) Manganese and cobalt binding in a multi-histidinic fragment. Dalton Trans 42:16293–16301
Pollmann K, Kutschke S, Matys S, Kostudis S, Hopfe S, Raff J (2016) novel biotechnological approaches for the recovery of metals from primary and secondary resources. Minerals 6:54
Qu W, Xue Y, Ding Q (2015) Display of fungi xylanase on Escherichia coli cell surface and use of the enzyme in xylan biodegradation. Curr Microbiol 70:779–785
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324
Samuelson P, Gunneriusson E, Nygren P-Å, Ståhl S (2002) Display of proteins on bacteria. J Biotechnol 96:129–154
Schippers A, Hedrich S, Vasters J, Drobe M, Sand W, Willscher S (2014) Biomining: metal recovery from ores with microorganisms. Adv Biochem Eng Biotechnol 141:1–47
Tsai D-Y, Tsai Y-J, Yen C-H, Ouyang C-Y, Yeh Y-C (2015) Bacterial surface display of metal binding peptides as whole-cell biocatalysts for 4-nitroaniline reduction. RSC Adv 5:87998–88001
Van Bloois E, Winter RT, Kolmar H, Fraaije MW (2011) Decorating microbes: surface display of proteins on Escherichia coli. Trends Microbiol 29:79–86
Wernérus H, Ståhl S (2004) Biotechnological applications for surface-engineered bacteria. Biotechnol Appl Biochem 40:209–228
Wu CH, Mulchandani A, Chen W (2008) Versatile microbial surface-display for environmental remediation and biofuels production. Trends Microbiol 16:181–188
Xu Z, Lee SY (1999) Display of polyhistidine peptides on the Escherichia coli cell surface by using outer membrane protein C as an anchoring motif. Appl Environ Microbiol 65:5142–5147
Yim S, An S, Han M-J, Choi J, Jeong K (2013) Isolation of a potential anchoring motif based on proteome analysis of Escherichia coli and its use for cell surface display. Appl Biochem Biotechnol 170:787–804
Acknowledgements
This work was supported by a grant from the Next-Generation BioGreen 21 Program (SSAC, Grant Number: PJ01111601), Rural Development Administration, Republic of Korea, and Research and Development Program of the Korea Institute of Energy Research (KIER) (B7-2437-01).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None of the authors declares a conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Maruthamuthu, M.k., Selvamani, V., Nadarajan, S.P. et al. Manganese and cobalt recovery by surface display of metal binding peptide on various loops of OmpC in Escherichia coli . J Ind Microbiol Biotechnol 45, 31–41 (2018). https://doi.org/10.1007/s10295-017-1989-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-017-1989-x