Abstract
Removal of 3-nitro-1,2,4-triazol-5-one (NTO) was investigated in conjunction with heterotrophic and autotrophic denitrifying growth conditions by a microbial consortium from a wastewater treatment plant. Microcosms were supplemented with molasses, methanol, or thiosulfate. Cultures were passaged twice by transferring 10 % of the culture volume to fresh media on days 11 and 21. Rates of NTO removal were 18.71 ± 0.65, 9.04 ± 2.61, and 4.34 ± 2.72 mg/L/day while rates of nitrate removal were 20.08 ± 1.13, 21.58 ± 1.20, and 24.84 ± 1.26 mg/L/day, respectively, for molasses, methanol, or thiosulfate. Metagenomic analysis showed that Proteobacteria and Firmicutes were the major phyla in the microbial communities. In molasses supplemented cultures, the community profile at the family level changed over time with Pseudomonadaceae the most abundant (67.4 %) at day 11, Clostridiaceae (65.7 %) at day 21, and Sporolactobacillaceae (35.4 %) and Clostridiaceae (41.0 %) at day 29. Pseudomonadaceae was the dominant family in methanol and thiosulfate supplemented cultures from day 21 to 29 with 76.6 and 81.6 % relative abundance, respectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Industrial wastewaters from fertilizer and munitions manufacturing as well as the mineral and metal processing industry are of concern because low organic carbon inputs limit the amount of biological nitrate removal [1]. For example, nitrate levels in explosives manufacturing wastewater can be as high as 3 g/L and are often accompanied by other toxic organic compounds [2]. As a result, these wastewaters often have a very low pH since the nitrates are typically discharged as nitric acid [1]. The removal of nitrates along with other nutrients from wastewater is ecologically important to prevent downstream eutrophication of lakes and rivers which can contribute to the formation of toxic algal blooms [3]. Nitrate is the most common nonpoint source contaminant in groundwater [4] and the United States Environmental Protection Agency (EPA) has established a maximum limit of nitrate in drinking water at 10 mg/L [5].
The biological dissimilatory transformation of nitrite or nitrate to N2 under anoxic conditions, where N oxide species serve as the terminal electron acceptor instead of O2 [6], is a key step in the cycling of nitrogen in the biosphere. Autotrophic denitrifiers are able to use inorganic sources such as reduced sulfur, ferrous iron, or H2 as electron donors [6, 7] to transform nitrate and nitrite. Autotrophic denitrification in the context of wastewater treatment has several potential advantages including lower costs, since no external organic carbon is needed, and lower sludge production [4, 8]. Sulfur based autotrophic denitrification is a particularly attractive approach because of the low cost and availability of elemental sulfur [5]. Several members of the α, β, γ, and ε-Proteobacteria are able to carry out autotrophic denitrification with 14 species being described to date [6]. In addition, some bacterial species capable of only reducing nitrate to nitrite with sulfur as an electron donor have also been identified [6]. In contrast to autotrophic denitrification, heterotrophic denitrification utilizes organic carbon as the electron donor while nitrate acts as the electron acceptor. Heterotrophic denitrification has been studied with a variety of simple and complex carbon sources including ethanol, methanol, propionate, acetic acid, sucrose, molasses, cotton, wheat straw, and polyhydroxyalkanoates (PHAs) [1, 9–11]. A variety of Gram-negative bacteria including the Pseudomonas, Thiobacillus, Alcaligenes, and Paracoccus genera as well as Gram-positive bacteria such as the Bacillus are able to perform heterotrophic denitrification [3].
While wastewater denitrification has been extensively studied, much less is known regarding the potential for biodegradation of nitroaromatic compounds in industrial wastewaters [12]. Biodegradation of energetic materials is an attractive remediation strategy, however, the breakdown of these compounds release additional nitrite into the wastewater effluent [13]. Therefore, biological processes that utilize microorganisms able to remove both nitroaromatics and nitrates would be an optimal treatment strategy for a wastewater environment.
Nitroaromatics are common constituents in many explosives, including modern insensitive munitions such as IMX-101 and IMX-104. These munitions contain known toxic aromatics such as hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) as well as lesser characterized compounds like 3-nitro-1,2,4-triazol-5-one (NTO), 2,4-dinitroanisole (DNAN), and nitroguanidine (NQ). While the biological processes for RDX degradation have been extensively studied, less is known about NTO biodegradation [13–15] Industrial production of munitions such as NTO leads to the contamination of large quantities of water with NTO and the commensurate biotic and abiotic breakdown products [16, 17]. Although considered less toxic than legacy explosives, NTO is highly soluble and thus of considerable environmental concern [18]. Biological reduction of NTO has been demonstrated in only a few cases [16, 18, 19] and more work is needed to identify the microorganisms responsible for NTO degradation and their associated biodegradation mechanisms. Further efforts are also necessary to determine the capacity of a microbial consortia in wastewater sludge to degrade NTO in conjunction with denitrification and to compare the effectiveness of heterotrophic and autotrophic growth conditions in the removal of both NTO and nitrates.
Here we compare the transformation of NTO and commensurate removal of nitrates under heterotrophic and autotrophic denitrifying conditions in batch reactors with enrichment cultures obtained from wastewater treatment plant (WWTP) sludge. Changes in community profile were investigated over the course of the experiment to infer key microbial phylotypes that correlate with nitrate removal and explosives transformations.
Materials and methods
Culture conditions
Batch studies were performed to assess the effectiveness of various organic and inorganic compounds on denitrification and NTO transformation rates. Microcosms were prepared in 150 mL serum bottles capped with rubber septa. Cultures were grown in M9 medium [20], consisting of a modified base nutrient basal salts solution containing 100 mg/L NTO, 3 g/L KH2PO4, 6 g/L Na2HPO4, 0.5 g/L NaCl, 1 mg/L MgCl2·6H2O, 1 mg/L MgSO4, 1 mg/L CaCl2, and 1 mg/L FeSO4. A M9 medium stock salt solution was prepared following Maniatis et al. [20], with the exception that CoCl2·6H2O was replaced with Co(NO3)2·6H2O. The medium was inoculated with 1 % anaerobic digester liquor (vol:vol) obtained from a municipal Wastewater Treatment Plant (Vicksburg, MS, USA). For heterotrophic denitrification, microcosms were supplemented with 1 % (vol:vol) molasses or methanol. Autotrophic denitrifying microcosms were supplemented with 28 mM thiosulfate. The headspace was flushed with N2 gas for 5 min and the serum bottles were incubated at room temperature with shaking at 100 rpm in the dark. On days 11 and 21, 10 % of the culture volume was transferred into new serum bottles containing fresh media, NTO, and carbon or thiosulfate at the same concentrations as the initial inoculation. Control microcosms were prepared as above and autoclaved three times prior to incubation. No growth was observed in autoclaved controls. All experiments were performed in triplicate.
Analytical methods
NTO, nitrite, and nitrate concentrations were measured every 3–4 days. NTO concentrations were measured by first filtering a 1 mL sample taken from each microcosm through a 0.45 μm PTFE filter. Samples were analyzed following a method modified from [18] on an Agilent 1100 HPLC equipped with a 100 mm × 3 mm, 5 μm particle size Hypercarb porous graphitic column (Thermo Scientific, Sunnyvale, CA, USA) with a Phenomenex Security Guard C18 analytical graphitic guard column (Phenomenex, Torrance, CA, USA). The elution was performed with a gradient mobile phase as follows: 0–3 min; 0.05 % trifluoroacetic acid (TFA) in water, 3–11 min; 15 % acetonitrile (ACN): 85 %, 0.05 % TFA, 11–15 min; 50 % ACN: 50 %, 0.05 % TFA, 17–24 min; 0.05 % TFA. The flow rate was 1 mL/min with a column temperature of 32 °C and a UV–Vis DAD at 315 nm. Nitrite, nitrate, and sulfate concentrations were measured on a Dionex ICS-3000 (Thermo Scientific, Sunnyvale, CA, USA) equipped with an IonPac AS20 column with conditions as specified by the column manufacturer (Thermo Scientific, USA). Briefly, the conditions were a 10 mM KOH eluent concentration and 1 mL/min flow rate, with a suppressor current of13 mA and a column temperature of 30 °C.
Metagenomic analysis
Samples were collected from the microcosms on days 11, 21, and 29 and from the initial WWTP sludge inoculum for 16S rRNA sequencing. Genomic DNA was extracted from the microcosms using the MoBio PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA) according to the manufacturer’s instructions. Following the protocol of [21], a 16S rRNA amplicon was generated using the primer pair 515F–806R with each reverse primer containing a barcode. Amplicons were then purified using a Qiaquick spin column (Qiagen, Valencia, CA, USA), quantified, and sequenced on an Illumina MiSeq (Illumina, San Diego, CA, USA) sequencer using the Illumina MiSeq Reagent Kit v3 (300 cycles) following the manufacturer’s instructions.
The 16S rRNA sequencing data from the Illumina runs were trimmed, demultiplexed and quality filtered with Illumina MiSeq Reporter. OTU assignment was performed with the open reference OTU clustering package implemented in the Quantitative Insights Into Microbial Ecology (QIIME) bioinformatics pipeline [22]. This workflow performs an initial closed reference clustering against the Greengenes database. Sequences that failed to hit the database were subsampled then clustered de novo and each cluster centroid was used as a reference sequence for a subsequent round of clustering against the Greengenes database. Sequences that failed to hit the database were subjected to a final round of de novo clustering.
Alpha diversity analyses were performed in QIIME using PD whole tree and observed species indices while beta diversity analyses were performed using weighted UniFrac [23] distance metrics. Principal coordinates analysis (PCoA) and PERMANOVA statistical analyses were also performed in QIIME. PERMANOVA tests were performed using 1000 permutations to estimate P values for differences between treatments. Beta significance between samples was computed using the weighted normalized UniFrac distances. Box plots of the PD whole tree alpha diversity were generated with GraphPad Prism 6 for Mac (GraphPad Software, La Jolla, CA, USA, http://www.graphpad.com) and 2D PCoA and rarefaction plots were generated using QIIME.
Results
Microcosm experiments
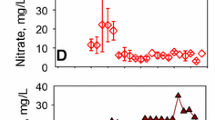
Microcosms supplemented with molasses were able to completely degrade 100 mg/L NTO by day 11 (Fig. 1a). In contrast, methanol and thiosulfate supplemented cultures had 40 ± 2.6 and 55 ± 6.4 mg/L NTO remaining, respectively (Fig. 1b, c). Following the first dilution of the cultures into fresh media on day 11, the methanol subcultures were able to completely degrade NTO by day 21 while the thiosulfate supplemented cultures still contained 42.8 ± 10.5 mg/L NTO. Following the second dilution of the cultures on day 21, NTO was degraded more slowly in the cultures with molasses with only 23.6 ± 4.2 mg/L remaining by day 29. The microcosms supplemented with methanol and thiosulfate showed no degradation of NTO between day 21 and 29. Overall, the highest rates of NTO degradation occurred following the second inoculation on day 11. Molasses supplemented cultures removed 18.71 ± 0.65 mg/L day while methanol and thiosulfate supplemented cultures removed 9.04 ± 2.61 and 4.34 ± 2.72 mg/L day, respectively (Table 1).
Concentrations of NTO, nitrite, and nitrate over time in waste water microcosms amended with molasses (a), methanol (b), and thiosulfate (c) compared with increases in the predominant families in each treatment. Specific families enriched in each microcosm are shown as percent relative abundance (left axis). Arrows indicate transfer of 10 % of the culture to fresh media with 100 ppm NTO on day 11 and 21. Error bars represent SD of three replicates
The initial concentration of nitrate was 50 mg/L and was rapidly degraded to less than 5 mg/L by day 4 in molasses and methanol supplemented cultures (Fig. 1a, b). In contrast, microcosms with thiosulfate had very little nitrate degradation by day 4 (Fig. 1c). Nitrate removal rates were also calculated under all treatment conditions. Following the initial inoculation, cultures supplemented with molasses and methanol had nitrate removal rates of 13.69 ± 1.03 and 13.47 ± 1.16 mg/L day, respectively, while the rate of nitrogen removal in cultures supplemented with thiosulfate was only 5.40 ± 5.69 mg/L day (Table 1). Similar nitrate uptake rates were observed following the second inoculation (Table 1). There was no measurable nitrite in the molasses supplemented microcosms at any point in the experiment. In contrast, nitrite increased from 0 to 44 mg/L in the methanol supplemented culture by day 4, but was no longer detectable by day 8 (Fig. 2a). Sulfate concentrations were also monitored throughout the experiment in the autotrophic denitrifying microcosms to verify that thiosulfate was being used as an electron donor. The initial sulfate concentration was around 300 mg/L (Fig. 2b) and it increased to 1,700 mg/L by day 21.
Metagenomic analysis
PD whole tree rarefaction curves were generated at a depth of 60,000 reads per sample. This was a sufficient sampling depth to represent the OTU richness of the microcosms (Fig. S1). Phylogenetic diversity metrics of microbial richness showed a significant decrease (P < 0.001) in PD whole tree alpha diversity (Faith’s phylogenetic diversity) in the microcosms between day 0 and day 11 of the experiment (Fig. 3). Alpha diversity did not change significantly between day 11 and day 29 in the molasses supplemented cultures. In contrast, alpha diversity decreased more gradually over time in microcosms supplemented with methanol and thiosulfate between days 11 and 29. Overall, there was no statistically significant difference in the PD whole tree alpha diversity, which is based on Faith’s phylogenetic diversity and observed OTUs, which is a non-phylogenetic diversity metric, between the different supplements by day 29.
Microbial community structure (beta diversity) was also compared between the different treatments using PCoA of the weighted UniFrac distances. The microcosms supplemented with molasses formed a distinct cluster (PERMANOVA, P < 0.001) from cultures supplemented with methanol and thiosulfate (Fig. 4). The carbon source accounted for 15.95 % of the variation between populations indicating that molasses had the greatest effect on community diversity.
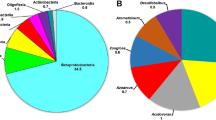
The relative abundance of taxa was compared on the family level to identify organisms that were able to take advantage of each of the carbon sources (Fig. 5). Of the dominant families (relative abundance >1 %), 15 had significant differences in abundance [false discovery rate (FDR) corrected ANOVA test: P < 0.05] (Table 2). All of the families belonged to the Proteobacteria, Firmicutes, or Bacteroidetes Phyla (Table 2).
To identify organisms that could potentially contribute to nitrogen removal in the microcosms, the abundance of individual families was compared to NTO, nitrite, and nitrate concentrations (Figs. 1a, 2a). Members of the family Pseudomonadaceae were at only 0.46 % relative abundance on day 0 but by day 11 represented 67.4 ± 5.5 % relative abundance in molasses supplemented microcosms (Figs. 1a, 5). This increase corresponded to a complete removal of NTO and nitrate over the same time period (Fig. 1a). Following dilution in fresh media on day 11, the relative abundance of Pseudomonadaceae decreased to 3.8 ± 0.6 % on day 21 and represented only around 1 % by day 29. Clostridiaceae also increased significantly but at a slower rate than Pseudomonadaceae. By day 11 Clostridiaceae was at 25.0 ± 6.4 % relative abundance and increased to 65.7 ± 17.9 % by day 21. Hydrogenophilaceae represented 33.42 ± 0.18 % relative abundance by day 11 in thiosulfate cultures and 23.7 ± 0.21 % in day 29 methanol cultures (Table 2). Closer analysis of this family showed that this increase in abundance was entirely attributed to the genus Thiobacillus. The family Bacillaceae also increased significantly in molasses supplemented cultures and reached a maximum of 12.07 % relative abundance by day 29 (Table 2). Subsequent analysis showed that this increase was due predominately to the genus Bacillus.
Discussion
The results of this study demonstrated that NTO can be efficiently removed in conjunction with either heterotrophic or autotrophic denitrification. Heterotrophic microcosms had higher rates of both NTO and nitrate removal than autotrophic microcosms. A comparison of heterotrophic microcosms showed that removal rates of NTO were significantly higher in cultures supplemented with molasses than in those fed methanol. This is not surprising given that molasses is a very rich, complex carbon source and was likely utilized by multiple genera. In addition, some organisms cannot use methanol as a sole carbon source which may have limited NTO and nitrate removal in those microcosms [24]. The declining rates of NTO degradation with successive inoculation may indicate a change to methanogenic degradation of NTO, since the nitrate was rapidly utilized with each successive inoculation. Methanogenesis is a slow process, in general, and slower rates of RDX degradation coupled to methanogenesis have been observed [25]. Similarly, denitrification coupled to thiosulfate oxidation generates acidic conditions which could negatively affect the microbial population and slow degradation rates in autotrophic microcosms [8].
Low levels of nitrite remained in the autotrophic microcosms on day 29 (Fig. 2a). Previous work has shown that cultures using sulfur for autotrophic denitrification could not utilize nitrate and nitrite together which resulted in high levels of nitrite accumulation [26]. The authors also found that the addition of a carbon source resulted in a decrease in nitrite accumulation. The fact that nitrite levels were low in cultures supplemented with thiosulfate indicates that some heterotrophic denitrification was occurring. It has been suggested that some heterotrophic activity can occur by utilizing organic carbon from dead biomass [26]. While some heterotrophic denitrification may have occurred, the significant increase in sulfate levels (Fig. 2b) provides additional evidence that autotrophic denitrification was primarily responsible for nitrate removal in these microcosms.
The significant decrease in alpha diversity by day 11 in molasses supplemented microcosms indicates that molasses, as a rich complex carbon source, enabled rapid growth which led to the population diversity stabilizing more rapidly. By day 29 all of the microcosms had similar alpha diversity with around 500 observed OTUs (Fig. 3b). This OTU richness is consistent with other reports for wastewater treatment sludge [27].
Metagenomic sequencing implicated several families of organisms in the removal of nitrogen. Members of phylum Proteobacteria were the predominant organisms under all treatment conditions while Pseudomonadaceae was the most abundant family (Fig. 5). This is consistent with other reports of Proteobacteria being the predominant organisms in soils and activated sludge [28, 29]. While members of the Pseudomonadaceae family are among the most abundant soil microorganisms [30], multiple species have also been implicated in denitrification, including Pseudomonas sp. C27, P. stutzeri, P. aeruginosa, P. aerogenes, P. chlororaphis, P. aureofaciens and P. caeni sp. [31–35]. They are known to use primarily nitrate as an electron acceptor and some species are capable of catalyzing the complete conversion of nitrite to N2 [36]. The increase in the relative abundance of Pseudomonadaceae with the corresponding disappearance of NTO and nitrate indicates that species from this family likely played a dominant role in heterotrophic denitrification. The loss of Pseudomonadaceae and the commensurate rise of Clostridiaceae in microcosms supplemented with molasses suggest that although members of the Clostridiaceae family are slower growing they may have a competitive advantage in the long term.
Several families of the β-Proteobacteria within the order Methylophilales were also significantly enriched by day 11 in the methanol and thiosulfate supplemented cultures (Table 2). Methylophilaceae has been shown previously to perform heterotrophic denitrification while oxidizing methanol [37]. In contrast, the increase in abundance of the genus Thiobacillus in the presence of thiosulfate indicated autotrophic denitrification. Thiobacillus denitrificans has been used in autotrophic denitrification of industrial wastewaters using sulfate as an electron donor [38]. The increase of this genus by day 11 in cultures with thiosulfate indicates that it may have been predominately responsible for denitrification between day 0 and 11 (Fig. 1c).
The increase in abundance of the Bacillaceae is also indicative of denitrification, as numerous Bacillus strains are primarily associated with aerobic heterotrophic denitrification [39]. Multiple species of the genus Bacillus have been implicated in denitrification and are commonly found in soils and wastewater, however their role in this process is not completely understood [40]. Anaerobic growth of several Bacillus strains in the presence of nitrate has been demonstrated [41, 42] as has in the aerobic reduction of NTO to 3-amino-1,2,4-triazol-5-one (ATO) [16]. Although nitrate levels were comparable under all treatment conditions, Bacillaceae was only observed in molasses fed cultures, which contained multiple complex sugars, further supporting the idea of anaerobic growth with nitrate as a terminal electron acceptor [42].
Conclusion
We demonstrated the removal of NTO under both autotrophic and heterotrophic denitrifying conditions. Rates of both NTO and nitrate removal were highest under heterotrophic conditions. Metagenomic sequencing showed that Proteobacteria was the dominant phylum while Pseudomonadaceae was the most abundant family. The enrichment of families containing known denitrifiers coupled with the subsequent removal of NTO and nitrate indicates that supplementing wastewater with a low cost carbon source may be an effective means of removing the insensitive munition NTO in conjunction with denitrification.
References
De Filippis P, Di Palma L, Scarsella M, Verdone N (2013) Biological denitrification of high-nitrate wastewaters: a comparison between three electron donors. Chem Eng 32:319–324
Cyplik P, Marecik R, Piotrowska-Cyplik A et al (2012) Biological denitrification of high nitrate processing wastewaters from explosives production plant. Water Air Soil Pollut 223:1791–1800. doi:10.1007/s11270-011-0984-5
Breisha GZ, Winter J (2010) Bio-removal of nitrogen from wastewaters—a review. J Am Sci 6:508–528
Pu J, Feng C, Liu Y et al (2014) Pyrite-based autotrophic denitrification for remediation of nitrate contaminated groundwater. Bioresour Technol 173:117–123. doi:10.1016/j.biortech.2014.09.092
Demirel S, Bayhan I (2013) Nitrate and bromate removal by autotrophic and heterotrophic denitrification processes: batch experiments. J Environ Health Sci Eng 11:27. doi:10.1186/2052-336X-11-27
Shao M-F, Zhang T, Fang HH-P (2010) Sulfur-driven autotrophic denitrification: diversity, biochemistry, and engineering applications. Appl Microbiol Biotechnol 88:1027–1042. doi:10.1007/s00253-010-2847-1
Wang R, Zheng P, Xing Y-J et al (2014) Anaerobic ferrous oxidation by heterotrophic denitrifying enriched culture. J Ind Microbiol Biotechnol 41:803–809. doi:10.1007/s10295-014-1424-5
Zhang TC, Lampe DG (1999) Sulfur:limestone autotrophic denitrification processes for treatment of nitrate-contaminated water: batch experiments. Water Res 33:599–608. doi:10.1016/S0043-1354(98)00281-4
Constantin H, Fick M (1997) Influence of C-sources on the denitrification rate of a high-nitrate concentrated industrial wastewater. Water Res 31:583–589. doi:10.1016/S0043-1354(96)00268-0
Oh SE, Bum MS, Yoo YB, Zubair A (2003) Nitrate removal by simultaneous sulfur utilizing autotrophic and heterotrophic denitrification under different organics and alkalinity conditions: batch experiments. Water Sci Technol 47:237–244
Wang X, Wang J (2009) Removal of nitrate from groundwater by heterotrophic denitrification using the solid carbon source. Sci China Ser B Chem 52:236–240. doi:10.1007/s11426-008-0111-7
Khalid A, Arshad M, Crowley DE (2009) Biodegradation potential of pure and mixed bacterial cultures for removal of 4-nitroaniline from textile dye wastewater. Water Res 43:1110–1116. doi:10.1016/j.watres.2008.11.045
Crocker F, Indest K (2006) Biodegradation of the cyclic nitramine explosives RDX, HMX, and CL-20. Appl Microbiol Biotechnol 73:274–290
Indest KJ, Hancock DE, Jung CM et al (2013) Role of nitrogen limitation in transformation of RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine) by Gordonia sp. strain KTR9. Appl Environ Microbiol 79:1746–1750. doi:10.1128/AEM.03905-12
Indest KJ, Jung CM, Chen H-P et al (2010) Functional characterization of pGKT2, a 182-kilobase plasmid containing the xplAB genes, which are involved in the degradation of hexahydro-1,3,5-trinitro-1,3,5-triazine by Gordonia sp. strain KTR9. Appl Environ Microbiol 76:6329–6337. doi:10.1128/AEM.01217-10
Le Campion L, Vandais A, Ouazzani J (1999) Microbial remediation of NTO in aqueous industrial wastes. FEMS Microbiol Lett 176:197–203
Richard T, Weidhaas J (2014) Biodegradation of IMX-101 explosive formulation constituents: 2,4-dinitroanisole (DNAN), 3-nitro-1,2,4-triazol-5-one (NTO), and nitroguanidine. J Hazard Mater 280:372–379. doi:10.1016/j.jhazmat.2014.08.019
Krzmarzick MJ, Khatiwada R, Olivares CI et al (2015) Biotransformation and degradation of the insensitive munitions compound, 3-nitro-1,2,4-triazol-5-one, by soil bacterial communities. Environ Sci Technol 49:5681–5688. doi:10.1021/acs.est.5b00511
Sarlauskas J, Nemeikaite-Ceniene A, Anusevicius Z et al (2004) Enzymatic redox properties of novel nitrotriazole explosives implications for their toxicity. Z Naturforsch C 59:399–404
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual, 1st edn. Cold Spring Harbor Laboratory Press, NY
Costello EK, Lauber CL, Hamady M, Fierer N (2009) Bacterial community variation in human body habitats across space and time. Science. doi:10.1126/science.1180060
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi:10.1038/nmeth.f.303
Lozupone C, Lladser ME, Knights D et al (2010) UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172. doi:10.1038/ismej.2010.133
Eberly JO, Ringelberg DB, Indest KJ (2013) Physiological characterization of lipid accumulation and in vivo ester formation in Gordonia sp. KTR9. J Ind Microbiol Biotechnol 40:201–208. doi:10.1007/s10295-012-1218-6
Waisner S, Fredrickson H, Hansen L, Barnerji K (2000) Removal of RDX from a contaminated groundwater by in situ bioremediation. USACE ERDC/EL Tech Report TR-00-14
Qambrani NA, Jung SH, Ok YS et al (2013) Nitrate-contaminated groundwater remediation by combined autotrophic and heterotrophic denitrification for sulfate and pH control: batch tests. Environ Sci Pollut Res 20:9084–9091. doi:10.1007/s11356-013-1623-z
Liao R, Shen K, Li A-M et al (2013) High-nitrate wastewater treatment in an expanded granular sludge bed reactor and microbial diversity using 454 pyrosequencing analysis. Bioresour Technol 134:190–197. doi:10.1016/j.biortech.2012.12.057
Roesch LFW, Fulthorpe RR, Riva A et al (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1–8. doi:10.1038/ismej.2007.53
Hu M, Wang X, Wen X, Xia Y (2012) Microbial community structures in different wastewater treatment plants as revealed by 454-pyrosequencing analysis. Bioresour Technol 117:72–79. doi:10.1016/j.biortech.2012.04.061
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728. doi:10.1128/AEM.72.3.1719-1728.2006
Knowles R (1982) Denitrification. Microbiol Rev 46:43–70
Guo H, Chen C, Lee D-J et al (2014) Proteomic analysis of sulfur–nitrogen–carbon removal by Pseudomonas sp. C27 under micro-aeration condition. Enzyme Microb Technol 56:20–27. doi:10.1016/j.enzmictec.2013.12.013
Lalucat J, Bennasar A, Bosch R et al (2006) Biology of Pseudomonas stutzeri. Microbiol Mol Biol Rev 70:510–547. doi:10.1128/MMBR.00047-05
Kathiravan V, Krishnani KK (2013) Pseudomonas aeruginosa and Achromobacter sp.: nitrifying aerobic denitrifiers have a plasmid encoding for denitrifying functional genes. World J Microbiol Biotechnol 30:1187–1198. doi:10.1007/s11274-013-1543-6
Xiao YP, Hui W, Wang Q et al (2009) Pseudomonas caeni sp. nov., a denitrifying bacterium isolated from the sludge of an anaerobic ammonium-oxidizing bioreactor. Int J Syst Evol Microbiol 59:2594–2598. doi:10.1099/ijs.0.005108-0
Schreiber K, Krieger R, Benkert B et al (2007) The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J Bacteriol 189:4310–4314. doi:10.1128/JB.00240-07
Kalyuhznaya MG, Martens Habbena W, Wang T et al (2009) Methylophilaceae link methanol oxidation to denitrification in freshwater lake sediment as suggested by stable isotope probing and pure culture analysis. Environ Microbiol Rep 1:385–392. doi:10.1111/j.1758-2229.2009.00046.x
Claus G, Kutzner HJ (1985) Physiology and kinetics of autotrophic denitrification by Thiobacillus denitrificans. Appl Microbiol Biotechnol 22:283–288. doi:10.1007/BF00252031
Kim JK, Park KJ, Cho KS et al (2005) Aerobic nitrification–denitrification by heterotrophic Bacillus strains. Bioresour Technol 96:1897–1906. doi:10.1016/j.biortech.2005.01.040
Verbaendert I, De Vos P (2011) Studying denitrification by aerobic endospore-forming bacteria in soil. In: Logan NA, Vos P (eds) Endospore-forming soil bacteria. Springer Berlin Heidelberg, Berlin, pp 271–285
Schirawski J, Unden G (1995) Anaerobic respiration of Bacillus macerans with fumarate, TMAO, nitrate and nitrite and regulation of the pathways by oxygen and nitrate. Arch Microbiol 163:148–154. doi:10.1007/BF00381790
Nakano MM, Dailly YP, Zuber P, Clark DP (1997) Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. J Bacteriol 179:6749–6755
Acknowledgments
This research was funded through the US Army Corps of Engineers Environmental Quality Program. Views, opinions and/or findings contained herein are those of the authors and should not be construed as an official Department of the Army position or decision, unless so designated by other official documentation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10295_2016_1755_MOESM1_ESM.png
Figure S1: PD Whole Tree rarefaction curves at a sampling depth of 60000 for each treatment. Red; day 0 control, Blue; methanol, Orange; molasses, Green; thiosulfate (PNG 35 kb)
Rights and permissions
About this article
Cite this article
Eberly, J.O., Indest, K.J., Hancock, D.E. et al. Metagenomic analysis of denitrifying wastewater enrichment cultures able to transform the explosive, 3-nitro-1,2,4-triazol-5-one (NTO). J Ind Microbiol Biotechnol 43, 795–805 (2016). https://doi.org/10.1007/s10295-016-1755-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1755-5