Abstract
Consumer wine preferences are changing rapidly towards exotic flavours and tastes. In this work, we tested five non-conventional yeast strains for their potential to improve Ribolla Gialla wine quality. These strains were previously selected from numerous yeasts interesting as food production candidates. Sequential fermentation of Ribolla Gialla grape juice with the addition of the Saccharomyces cerevisiae T73 Lalvin industrial strain was performed. Zygosaccharomyces kombuchaensis CBS8849 and Kazachstania gamospora CBS10400 demonstrated positive organoleptic properties and suitable fermentation dynamics, rapid sugar consumption and industrial strain compatibility. At the same time, Torulaspora microellipsoides CBS6641, Dekkera bruxellensis CBS2796 and Dekkera anomala CBS77 were unsuitable for wine production because of poor fermentation dynamics, inefficient sugar consumption and ethanol production levels and major organoleptic defects. Thus, we selected strains of K. gamospora and Z. kombuchaensis that significantly improved the usually plain taste of Ribolla wine by providing additional aromatic complexity in a controlled and reproducible manner.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Yeast fermentation of plant carbohydrate sources is one of the oldest human technologies dating back to the Neolithic era [11]. The yeast inoculum, however, remains one of the main focuses in wine research. Many attempts have been made to diversify aroma profiles in wine in both industrial and research enology sectors. The medieval, low-intervention and traditional spontaneous fermentation method remains the leading practice for obtaining the most complex and interesting wine profiles [4] because of the aromatic repertoire characteristic of the large number of yeast species involved. However, the complexity entailed with unknown natural microbiota on grape skins in vineyards and cellars makes it a risky and unpredictable practice. Reproducibility, easy monitoring, customer satisfaction and subsequent predictable business niches have led to the widespread use of single culture inocula in the modern wine industry. However, the main drawback of this type of fermentation is often the uniformly plain taste and flavour of the product [7, 17, 35], whereas spontaneous fermentation adds diversity in terms of wine characteristics.
The trend of pure starter cultures has drastically reduced the diversity of the yeast species employed in winemaking, and even single strains within species are used in wine fermentations. Although we know of thousands of different yeast species, only half a dozen of them are commercially available for winemakers [23]. In the search for new tastes, non-Saccharomyces strains were tested and some of them showed promising performance [3, 7, 14, 17, 18, 25, 46]. However, in the case of use of non-conventional yeasts or during spontaneous fermentations, problems may arise with reproducibility. Mixed-culture fermentation could be an attractive solution to enhance fermentation in a controlled manner. A recent trend for mixed-culture inoculations has already proven to be profitable for the quality of the final product in a number of studies [24, 36, 53]. This is why we used in our study the sequential inoculation approach to test five previously selected yeast strains for their potential to improve the aroma characteristics of Ribolla wine.
The Vitis vinifera L. cv. Ribolla Gialla is an autochthonous white grape variety cultivated in south-west Slovenia, north-eastern Italy and parts of Croatia. Since the wine made from this variety is mostly presented on the market as a table wine, the quality could be improved through various technological approaches. This is why our aim is to intensify the aroma profile of this beverage by modifying the inoculated yeast strains.
The tested yeast species were obtained following screening for organoleptic properties in the Cornucopia project. The EU ITN Cornucopia project (http://www.yeast-cornucopia.se) attempted to explore yeast biodiversity and screened hundreds of publically available yeast isolates (http://www.cbs.knaw.nl ) to identify new species with promising aroma potential that could be employed in wine fermentations, as well as beer brewing, cheese making and baking [37, 55]. This selection was based on their good performance in terms of aroma profile when lab scale fermentations were carried out. Kazachstania gamospora (CBS10400) and Torulaspora microellipsoides (CBS6641) excelled in the production of higher alcohols, ketones and certain aldehydes and acetate esters; Dekkera anomala (CBS77) and Zygosaccharomyces kombuchaensis (CBS8849) were remarkable in their production of certain higher alcohols and ethyl esters; and Dekkera bruxellensis (CBS2796) synthesised high quantity of certain higher alcohols.
In the current study, we evaluated the fermentative performance of D. anomala (CBS77), D. bruxellensis (CBS2796), K. gamospora (CBS10400), T. microellipsoides (CBS6641), Z. kombuchaensis (CBS8849) in successive fermentations with the industrial wine strain S. cerevisiae Lalvin T73. As the next step, selected K. gamospora and Z. kombuchaensis were tested in pilot-scale fermentations.
Materials and methods
Yeast strains and media
The yeast strains used in this study were obtained from the CBS yeast collection (Utrecht, The Netherlands, http://www.cbs.knaw.nl). They were: D. anomala (CBS77), D. bruxellensis (CBS2796), K. gamospora (CBS10400), T. microellipsoides (CBS6641) and Z. kombuchaensis (CBS8849), whereas the industrial wine strain S. cerevisiae Lalvin T73 was obtained from Institute of Agrochemistry and Food Technology in Valencia (Spain). More information about the origin of isolates is available in Table 1S.
All strains were selected from the CBS collection as they turned to be the most promising yeasts for food production. This screening was performed previously in the EU ITN Cornucopia project, in which species were tested for flavour production abilities in MicroBread, MicroBeer, MicroWine and MicroCheese microsystems [55].
Yeast Extract Peptone Dextrose rich media (YPD, Difco) was used for yeast cultivation and biomass production. For assessing the CFU (colony-forming unit) counts, mixed-culture fermentation samples were cultivated on Wallerstein Laboratory (WL) agar [54].
Grape must sequential fermentations
The yeast inoculum was prepared by growing the respective strains in YPD media at 25 °C with 200 rpm agitation. The overnight culture was harvested, washed with grape juice and left at room temperature in 40 mL of grape must until there were visible signs of fermentation (up to 3 h). Fermentation was performed in 500 mL Erlenmeyer flasks sealed with airlock lids. 1 × 106 cells/ml of the corresponding strain were inoculated into 400 mL of Ribolla Gialla grape juice. The frozen grape juice was adjusted to 20 °C (experimental temperatures) prior to inoculation. After 60 h of incubation the first samples were taken for determination of yeast viability using CFU counts and measurement of sugar and ethanol contents. In order to avoid oxidation, carbon dioxide was applied in the sampling area when opening the lids. Samples from the S. cerevisiae control were prepared as described above. The second and third sampling time points were performed after 150 and 300 h to assess CFU counts (using WL differential agar), ethanol and sugar contents. Additionally, samples from 300-h time points were taken for aroma profile analysis. Lastly, the rest of the sample was treated with 0.1 g/L of potassium bisulfite for tasting panel analysis [4, 48].
As the next step, we performed 5-L scaled-up fermentations with the K. gamospora, Z. kombuchaensis and S. cerevisiae as the control. Together with the increased fermentation volume we decreased the number of samplings in order to test fermentation behaviour in conditions more similar to those of the wine cellar. The fermentation time was also extended to 360 h. As a result, we obtained only final time point (360 h) samples form measurements of sugar and ethanol content and aroma profile analysis. The remaining amount of the sample was treated with 0.1 g/L of potassium bisulfite, filtered and bottled for tasting panel analysis.
Sugar, ethanol and weight loss measurements in wine must samples
Measurements of sugar and ethanol levels were assessed using the D-Fructose/D-Glucose Assay Kit and the Ethanol Assay Kit both from Megazymes (Ireland). Samples collected from the must fermentations in 400 mL scale were diluted according to the manufacturer’s instructions. The two main sugars in grape must, glucose and fructose, were measured using the D-Fructose/D-Glucose Assay Kit. In addition, the ethanol content was detected using an enzymatic Ethanol Assay Kit (Megazymes, Ireland). The reactions were mixed according to the manufacturer’s instructions and readings at 265 nm were obtained using a Perkin Elmer HTC5000 plate reader (Perkin Elmer, USA). The calculations were made using the data calculator provided by the manufacturer. In order to monitor the fermentation dynamics, weight loss reads due to the evolution of CO2 were obtained using a medium resolution balance (±0.1 g).
Batch cultivation in fermenters
Aerobic batch cultivation was performed in triplicate in Multifors (Infors HT, Bottingen, Switzerland) bioreactors, with a working volume of 1 L using minimal defined media [47]. Dissolved oxygen (monitored using an InPro 6800S sensor from Mettler Toledo, Switzerland) was maintained above 30 % using stirrers in cascade mode, varying the stirring speed between 200 and 1200 rpm, at 25 °C with airflow set at 1 L/min. The pH was maintained at 5 (±0.5) through automatic addition of 2 M KOH and 1 M H2SO4 and monitored with a 405-DPAS-SC-K8S/225pH sensor (Mettler Toledo, Switzerland). Gas analysers, BC-CO2 and BCP-O2 (Blue Sens, GmbH, Herten, Germany) were fitted on the exhaust lines to determine CO2 and O2 levels in the outflow, respectively. Seed cultures were grown in 100 mL of minimal medium in 500-mL baffled-bottom flasks for 16–24 h at 25 °C and 200 rpm, washed and used to inoculate all batch cultures as reported [47]. In order to test the fermenter culture for contamination, sequence analysis of LSU rDNA D1/D2 domains was applied. Sequencing was performed with primers NL1 (5′ GGTCCGTGTTTCAAGACGG 3′) and NL4 (5′ GCATATCAATAAGCGGAGGAAAAG 3′) [26, 50]. The results were aligned in the CBS identification database (http://www.cbs.knaw.nl/Collections/DefaultInfo.aspx?Page=Home) to confirm the identity of the strain of interest.
Growth kinetics and extracellular metabolite analysis
Cell density was monitored using both DW (dry weight) and OD (optical density) methods. The former was used to calculate metabolites production and glucose and oxygen consumption rates. Glass microfibre GF/A filters with a pore size of 0.45 μm (Whatman, Milan, Italy) were weighed before and after filtering of a known amount of sample, washed with distilled water, and then dried in an oven at 70 °C for a minimum of 24 h to determine cell dry weight. Weights of the filters containing dried cultures were assessed using the high-resolution balance (±0.0001 g). All growth kinetics were analysed with glucose as the sole carbon source. Cell growth was monitored in parallel by measuring OD600nm.
Samples taken during the exponential growth phase at appropriate intervals were centrifuged for 2 min at 16,000×g and the supernatant was filtered through a 0.2 μm membrane filter and then used to determine the concentration of glucose, ethanol, glycerol, pyruvate and organic acids using an HPLC 1200 series (Agilent, USA) equipped with a 300 × 7.7 mm Aminex HPX-87H ion exchange Column (Biorad, USA) set at 60 °C. 5 mM H2SO4 was used as a mobile phase at a flow rate of 0.6 mL × min−1. These compounds were detected using a refractive index detector (Agilent Technologies G1362A) set at 55 °C in series with a variable wavelength detector at 210 nm (Agilent Technologies G1314B). An Agilent ChemStation was used to calculate the area under the detected peaks, corresponding to the calibration curves of metabolite standards, performed using a multiple point calibration system with standards purchased from Sigma Aldrich.
Product yield was determined by computing the amount of product expressed on the basis of the amount of substrate consumed and presented as g/g. Specific glucose consumption rates (q glucose mmolg−1 × h−1) were calculated in the exponential growth phase, when glucose was the sole carbon source, by expressing the amount of glucose consumed on the basis of the biomass concentration increase rate (g × 1−1 × h−1), multiplied by the corresponding specific growth rate [μ max (h−1)] as reported before [22]. Similarly, when calculating the specific ethanol production rate (q ethanol mmolg−1 × h−1), the amount of glucose consumed per hour was replaced by the amount of ethanol produced per hour. Acetate production rates were calculated as described above. The respiratory quotient (RQ) was calculated as the ratio between carbon dioxide produced on the basis of the oxygen consumed. To verify the quality of each experiment, the carbon balance was computed.
Analysis of volatile profiles using GC–MS
Aromatic profiles were determined using a Thermo Scientific TSQ Quantum GC Triple Quadrupole GC/MS (Thermo Scientific, USA). The chromatographic column used was the VF-wax column, 30 m/ID 0.25 mm/Film 0.25 μm (Agilent Technologies, The Netherlands). The chromatograph was equipped with a PAL autosampler (PALsystem, Switzerland). In order to maximise the similarity to sensory analysis headspace, solid-phase micro-extraction (SPME) was adopted using Divinylbenzene/carboxen/polydimethylsiloxane fibre (DVB-CAR-PDMS, Supelco, USA). The method reported previously by Ravasio et al. [37] was partially modified in order to fit a specific matrix and potential metabolites.
Briefly, 1 mL of fermented must was added to 3.85 mL of deionised water, 2.2 g of sodium chloride, 50 µL of 0.1 % of sodium azide, 0.1 g of ascorbic acid and 2-octanol internal standard. The samples were incubated for 10 min at 40 °C. The headspace volatiles were then adsorbed on SPME fibre for 40 min and injected into the chromatographic system. Volatiles were identified using an internally created library, based on an in-house automatic processing system, described by Wehrens et al. [49] and the NIST mass spectral database (Nist 1.0.0.23). Quantification was carried out using ThermoExcalibursoftware (Version 1.0.1.03, Thermo scientific, USA). The compounds identified were expressed as internal standard equivalents using chromatographic peak ratios.
Data visualisation and statistical analysis
Data visualisation of volatile profiles was performed using a MultiExperiment viewer (TM4, USA) and the Statistica 9 package (StatSoft, USA). The data for different parameters, e.g. sugars, ethanol, CFU measurements, volatile profiles, were processed using one-way ANOVA (n.s.: not significant; *P < 0.05 (*); **P < 0.01; ***P < 0.001).
When ANOVA was significant, the means were separated using least significant difference (LSD) post hoc test (p < 0.05) using a Sigma Plot 13 (Systat Software, Inc., San Jose, CA). CFU data were subjected to logarithmic transformation before statistical analysis.
Tasting panel
Sensorial analysis was performed using technical triplicates according to the protocols outlined by Lawless and Heymann [29]. The main factors evaluated were colour, smell and taste. The evaluation panel was made up of eight people. Both trained experienced tasters and wine enthusiasts were represented in the group. Wine samples were served at 8 °C in tulip wine glasses. Samples were labelled with numeric codes and served in a randomised order. The first evaluation was based on the hedonic scale, in order to determine the most and least appreciated samples. Panellists were also asked to describe the attributes perceived in each sample. The second tasting session was performed in a more detailed way, by using the comparative descriptive profiling approach [43]. Each panellist was provided with a score-sheet with consensus vocabulary (attributes) with which the intensity of each attribute was rated using a linear unstructured scale, as previously described by Dairou and Sieffermann [10], however, with the attributes adjusted to white wine samples. The level of each attribute was calculated as the length of the segment marked by the taster. The more intensive is the perception of the attribute, the longer the fragment marked. After tasting was performed, each sample attribute was scored and subjected to ANOVA analysis. Tasters were considered as a random factor [31].
Results and discussion
Small-scale Ribolla fermentation
Sugar consumption, ethanol production and weight loss analysis
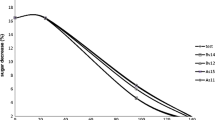
In order to evaluate whether the yeast species D. anomala (CBS77), D. bruxellensis (CBS2796), K. gamospora (CBS10400), T. microellipsoides (CBS6641) and Z. kombuchaensis (CBS8849), could be useful for the winemaking process, a small-scale experiment with 400 mL grape must was performed. After 60 h of fermentation with a single strain yeast culture, the industrial wine strain, S. cerevisae Lalvin T73 was added and fermentation continued for another 240 h. The sugar consumption rates were comparable for all five strains, but T. microellipsoides, D. bruxellensis and to a lesser extent D. anomala showed some delay in utilising sugar, especially fructose (Fig. 1). This can be explained by the increased glucose consumption in S. cerevisiae and K. gamospora, which are Crabtree-positive yeasts [12, 21]. Raw data for both sugar and ethanol measurements are available in Table 2S.
Sugar (a) and ethanol (b) concentrations measured after 60, 150 and 300 h of sequential fermentations with D. anomala (CBS77), D. bruxellensis (CBS2796), K. gamospora (CBS10400), T. microellipsoides (CBS6641), Z. kombuchaensis (CBS8849). First time point is representing metabolic activity of single strain after 60 h of fermentation, two other time points are measured when both strains are present. Statistical groups determined using LSD test P (< 0.05). Comparison was done in between corresponding timepoint values
The Dekkera lineage evolved the Crabtree effect independently of Saccharomyces [39]. Our experiment showed a slower disappearance of sugars in case of Dekkera species, as compared to the post-Whole Genome Duplication (WGD) genera Kazachstania and Saccharomyces [6, 27, 52]. Previous studies rated Dekkera species a good ethanol producer [5, 16, 40, 45]. The lower fermentative capacity observed could be explained by the Custers effect, i.e. inhibition of alcoholic fermentation during anaerobiosis [13]. As a confirmation of the partial redox imbalance in Dekkera, high levels of acetate in comparison to S. cerevisae were detected using GS–MS (Table 3S). It seems that despite the fact that Dekkera is a Crabtree-positive genus, high sugar concentrations cause a prolonged lag phase before the yeast cells start fermentation. After 300 h of fermentation there was still around 40 g of available sugar left (Fig. 1). It seems likely, that D. bruxellensis and D. anomala produced a larger amount of ethanol in the event that the experiment is prolonged.
The genus Zygosaccharomyces is well known for its preference for fructose over other sugars [42]. In our experiment, Z. kombuchaensis utilised fructose until depletion, while glucose levels were also reduced to 7–12 g/L (Fig. 1) depending on the experiment. This can be explained by the presence of the S. cerevisiae wine strain, which was added after 60 h, which stimulated the reduction of both sugars in wine must rapidly (Table 2S). Combination of S. cerevisiae and Z. kombuchaensis could be a good solution for wine production in case the dryness of the beverage cannot be obtained using only the Saccharomyces strain [20].
Ethanol levels detected varied from 25 to 70 g/L (Fig. 1). T. microellipsoides together with Dekkera species and Z. kombuchaensis, produced concentrations which were twice lower than those produced by S. cerevisiae and K. gamospora during wine must fermentation. As expected, elevated levels of ethanol produced by the post-WGD yeast (S. cerevisiae and K. gamospora) correlated with elevated sugar consumption levels. Both genera are characterised by an increased glycolytic flux due to the increased copy numbers of hexose transporters [32]. They have an enhanced ability to metabolise glucose and they are also Crabtree-positive, which means that ethanol is produced even when oxygen is present in the environment [8, 12, 33, 34, 51, 52].
Lastly, we observed a significant disappearance of sugars after three biological replicates involving fermentations with both K. gamospora and Z. kombuchaensis, prompting us to select them for further studies. The Dekkera and Torulaspora strains were eliminated for the pilot scale experiment, because of low fermentation dynamics and production of off flavours.
To complement our findings, we computed fermentation rates by measuring weight loss due to CO2 evolution. K. gamospora with Z. kombuchaensis, together with the industrial wine strain control showed a more dynamic weight loss, thus showing a desirable fermentation ability. In addition, the weight loss delay in Dekkera and T. microellipsoides wine fermentations also confirmed the findings (Fig. 1S).
Microbial analysis of fermentation samples
Longitudinal analysis of samples from wine must fermentations was done using viability tests based on estimation of CFU using differential Wallerstein agar [54]. Taking into consideration that successive inoculations of two different yeast strains were performed, samples from 150 and 300 h represented the metabolic activity of both strains. We therefore sought to confirm this assumption and verified the coexistence of both species in one flask during the experiment by plating out samples on WL agar and then estimating cell viability and the abundance of the two species (Table 1). In most cases, the yeast numbers reached the expected final values for a saturated culture at the end of the fermentation, from 1.7 × 105 for D. bruxellensis and up to 6.7 × 106 for S. cerevisiae. Surprisingly, in several cases S. cerevisiae could not complete growth and at the end of fermentation represented only a minority of the yeast population, for example in the case of inoculation with earlier Zygosaccharomyces kombuchaensis and Kazachstania gamospora. Thus, the two earlier inoculated yeasts were the main players in the fermentation process, and they somehow “inhibited” growth of S. cerevisiae, probably through production of lytic enzymes, sulphur dioxide, killer toxin peptides or partial depletion of nutrients or oxygenas was reported previously by Fleet [15]. In the case of Torulaspora and both Dekkera species, S. cerevisiae, reached 30 % of total biomass after inoculation, and all three yeasts produced higher amounts of acetic acid in comparison with S. cerevisae (Table 3S), which can affect its growth [41]. It is important to mention that Dekkera can even influence fermentation with very low cell counts [9]. Interaction between the Saccharomyces and Dekkera species during fermentation is now a pressing and elusive question. Because of the shared habitats, which are often industrial reactors, many researchers are trying to perform multifactorial analysis of the interaction between these two species. However, the interactions between representatives of Saccharomyces and Dekkera genera during fermentation are still little understood [2].
Aroma profile analysis
Using gas chromatography–mass spectrometry analysis the aroma profiles of tested yeast species were characterised. The end-point samples (300 h) of the small lab fermentations were analysed for their volatile profiles. Using the SPME–HS–GC–MS methodology 50 compounds were identified and associated with their aroma descriptors using The Good Scent Company database. (www.thegoodscentscompany.com). Data expressed as equivalents of internal standard using chromatographic peak ratios and the sensory descriptors of individual compounds are available in the supplementary material (Tables 3S and 4S, respectively). As relative concentrations do not allow comparison with literature they were used to characterise volatile profiles in comparison with S. cerevisae.
The total data set was visualised using a PCA plot with most influential loadings for sample separation (Fig. 2S).
According to the PCA plot, yeasts were grouped into three separate groups, with S. cerevisiae and Z. kombuchaensis forming the first group. The second group was made up of D. bruxellensis, D. anomala and T. microellipsoides and K. gamospora was separated from those two. The compounds that contributed most to separation of the first group were ethyl hexanoate, decanoic acid, 4-ethyl phenol, 4-ethyl guaiacol and 2-hydroxypropanoic ethanoate. The main compounds, contributing to the separation of K. gamospora were 2-phenyl ethanol, phenethylacetate, phenethyloctanoate and ethyl propionate, while isoamyl alcohol and 4-vinylguaiacol had the biggest influence on the separation of S. cerevisiae and Z. kombuchaensis (Fig. 2S).
The volatile profiles of both the Dekkera and Torulaspora species were made up by a large number of ethyl esters and higher alcohols, which could contribute to the fruity character of the wine produced. On the other hand, all three strains produced volatile phenols out of cinnamic acid. Expression of cinnamate decarboxylases and vinyl-phenol reductase could influence the wine produced by creating the “horse sweat” odour [19]. The aromatic profile described is tolerated to a certain extent in aged red wines, but is undesirable in white wine production, such as ours [38, 44]. Because of the high concentration of volatile phenols and negative sensory descriptors, D. bruxellensis, D. anomala, and T. microellipsoides were omitted from subsequent experimentation. Box-plot charts of mentioned compounds are presented in Fig. 2.
Boxplots of volatile compounds that are extensively contributing to distinguishing yeast aromatic profiles. DA D. anomala (CBS77), DB D. bruxellensis (CBS2796), KG K. gamospora (CBS10400), TM T. microellipsoides (CBS6641), ZK Z. kombuchaensis (CBS8849). Concentrations are expressed as IS equivalents using chromatographic peak area ratios. P and F values are calculated using one-way ANOVA
Kazachstania gamospora was characterised by the production of phenethyl alcohol, phenethyl acetate and ethyl propionate, which have the potential to contribute a fruity and flowery character which is not present in wines from the region. For this reason cofermentation with K. gamospora could improve the aromatic characteristics of this wine. The most similar characteristics to the control were observed after the fermentation of Ribolla Gialla with Z. kombuchaensis. The most different compounds, separating Z. kombuchaensis fermented samples from the samples fermented by the other tested species were isoamyl alcohol and 4-vinyl guaiacol. Both compounds are characteristic for fermentation with Saccharomyces strains [1]. The presence of 4-vinyl guaiacol shows expression of cinnamate decarboxylase, but not vinyl-phenol reductase and gives wine spicy and peppery notes, if higher than the sensory threshold (Fig. 2).
Tasting panel
On the basis of the results of the tasting panel using the hedonic scale, the group of D. anomala, D. bruxellensis and T. microellipsoides was evaluated as producing the least appreciated wines. D. anomala had high scores for colour evaluation; however, off-flavour compounds were detected both by GS–MS and sensorial analysis (Figs. 2, 3S). The off-flavours were mostly represented by volatile phenols formed from cinnamic acids, as can be seen from the chemical analysis of volatiles. Z. kombuchaensis and K. gamospora had high scores, considering colour, taste and smell attributes, as compared to wine samples made with S. cerevisiae, and were selected for the second round of sensory evaluation (Fig. 3S).
Radar chart of sensory attributes in the samples of young wines produced with K. gamospora (CBS10400)—diamond markers, Z. kombuchaensis (CBS8849)—triangle markers and S. cerevisiae Lalvin T73—squares, evaluated by eight panellists. Stars on the right top of the attribute are specifying the level of significance evaluated with one way ANOVA
In the second round of sensory evaluation, K. gamospora produced samples were characterised by the most flowery, honey-like, fresh aroma and the most persistent and intensive flavour (Fig. 3). This can be explained by the high concentrations of phenetyl alcohol, phenetyl acetate and ethyl propionate detected during the chemical analysis (Fig. 2; Table 3S). The fermentation product of K. gamospora was also evaluated as the most pleasant sample in terms of taste. Samples of Z. kombuchaensis fermentations were more similar to those formed by S. cerevisiae, but were given higher scores for the following attributes: intensity of flavour, quality of flavour, sweetish, honey-like and pleasant (not necessary wine-like) taste (Fig. 4S). Samples fermented with Z. kombuchaensis also had a more intensive flavour and in general the wine obtained was judged as being more complex. Raw data from sensory evaluation analysis can be accessed in Table 5S.
To summarise, both fermentation samples of K. gamospora and Z. kombuchaensis achieved high scores when compared to those resulting from S. cerevisiae only. The addition of non-conventional yeasts attributed interesting fruity notes, freshness and a smell of flowers and honey. The addition of non-conventional yeasts mostly had an impact on aroma attributes and to a lesser extent on colour and taste (Figs. 3, 4S). The wines produced with D. anomala, D. bruxellensis and T. microellipsoides were considered to be low-quality products with low sensorial qualities.
Pilot-scale fermentations were analysed in the same manner. Aroma, colour and taste attributes showed a similar distribution, with the exception of the scores obtained for K. gamospora-based fermentations. This time, the fruity flavour attribute decreased, which might be due to the extended fermentation time or changed growth rates.
Employing selected yeasts in Ribolla pilot-scale fermentation
There is a tendency for changing performance when fermentations are scaled up [30]. In order to test whether this was also true in our case, we carried out fermentations of Ribolla must in 5-L vessels. The same measurements were made, except that sampling was only performed after 360 h at the end of fermentation, to mimic the original winemaking process and avoid additional oxidation. The two selected yeast strains (K. gamospora CBS 10400 and Z. kombuchaensis CBS8849) with the S. cervisiae industrial strain, as the control, behaved in a similar way and some of the parameters even improved. For example, the ethanol levels and sugar consumption rates were higher as compared to the 400-mL scale experiment (Fig. 5S). This can be explained by minimal human intrusion in the fermentation process and more homogeneous conditions for the yeasts. Additional analysis of basic metabolites was obtained from HPLC measurements (Table 6S). After 360 h of fermentation, the samples still contained 4 % (S. cerevisiae) to 7 % (K. gamospora) of available sugars. Ethanol reached 7 % in all three cases. These data overall showed good fermentation potential for both strains tested.
The aromatic profiles from the large-scale fermentations are displayed as a heat map with Pearson correlation hierarchical clustering showing that volatiles clustered into four groups (Fig. 4). The group of acetic acid esters was more abundant in K. gamospora-based fermentations, while the group of ethyl esters was more significant in those by S. cerevisiae. In between there was a third group made up of higher fatty acids, such as octanoic, decanoic acid and isoamyl alcohol. These compounds were mostly equally distributed between the two strains. The fourth group was characteristic for Z. kombuchaensis-based fermentations and was made up of carbonyl compounds, such as hexanal and nonanal, acetaldehyde, furfural, and higher alcohols, such as hexanol, 2-ethyl hexanol, decanol, dodecanol and hexadecanol.
In a similar way to the small-scale fermentation experiment, there was a higher concentration of phenethyl alcohol and phenethyl acetate in the product made by K. gamospora in comparison with S. cerevisae. An additional contribution to wine aroma comes from the formation of different esters. Most acetic acid esters have a fruity character, which could be desirable in young white wines, although high concentrations can lead to the oxidised, (over) ripe, ethereal and sharp sensory descriptors common for ethyl acetate, acetaldehyde or amyl acetate [28]. Ethyl esters of higher fatty acids are characteristic of the fermentation by S. cerevisiae and do not contribute to an extensive fruity or flowery aroma. The sensory related descriptors are waxy and oily, with some fruity, flowery or green notes in the background.
In comparison to S. cerevisiae and K. gamospora, fermentations by Z. kombuchaensis resulted in low ester and medium chain fatty acid concentrations, but it was found to be slightly richer in higher alcohols, none of which are, however, known to be desirable aroma descriptors for young white wines. It is also possible that a long lag phase with low aroma and alcohol production contributed to the neutral character of the Z. kombuchaensis volatile profile.
Batch cultivation
In order to study the fermentation parameters of the two selected yeast strains with the industrial yeast S. cerevisiae more carefully, we performed batch cultivation in fully controlled aerobic fermenters using defined minimal media [47] for each of the strains separately. This made it possible to accurately analyse sugar consumption and ethanol production yields more carefully, as well as to determine other important intermediates accumulating during carbon metabolism. In this case, the strains were added as a pure culture, so that the individual metabolic pattern could be studied.
There was no significant difference between S. cerevisiae and K. gamospora in terms of their carbon utilisation (Fig. 5). The two species produced the same amounts of ethanol and biomass. However, K. gamospora produced about twice as much glycerol as compared to S. cerevisiae. In addition, acetate was not produced in fermenter conditions, but only in wine must fermentations. The specific production rates were also the same, except that K. gamospora exhibited a higher oxygen consumption rate (Fig. 6S).
Physiology of the K. gamospora (CBS10400), Z. kombuchaensis (CBS8849) and S. cerevisiae Lalvin T73 in fermenters under aerobic conditions in minimal defined medium. Data are shown as the mean of triplicate independent experiments ± SD. a Yields of metabolites (ethanol, acetate, glycerol, biomass) are shown in grams per gram of glucose consumed (g × g−1). Maximum specific growth rate [μmax(h−1)] of the three strains is also shown. b Specific glucose consumption rates (q glucose) are expressed as the amount of glucose consumed as a function of the rate of increase of biomass concentrations multiplied by the corresponding specific growth rate (mmol × g−1DW × h−1). Similarly, specific ethanol production rates (q ethanol) are expressed as the amount of the amount of ethanol produced per hour per gram of biomass. q acetate was calculated as above
The fermentative capacity of Z. kombuchaensis is too poor to be applicable in wine production as a single strain, but the results from the real wine must experiment showed that when combined with S. cerevisiae it performed well, resulting in lower alcohol levels and interesting aroma profiles (Figs. 1, 2). Furthermore, the strain seems to prefer respiration to fermentation, as shown by the lower respiratory capacity (RQ) (Fig. 6S). It is worth mentioning that both cultivations were done with glucose as the sole carbon source to make it possible to perform uniform experiments and calculations. Must, however, is a mixture of fructose and glucose, and species of Zygosaccharomyces prefer fructose over glucose [21, 42]. This can explain the better performance of the strain in wine must as compared to batch cultivation using minimal media.
Conclusions and perspectives
Grape must is a sugar-rich media without much taste and aroma. The role of yeasts in the conversion process to wine is very important. Sugars, in the form of glucose and fructose, must be depleted and ethanol produced. As side metabolites of the fermentation process numerous aroma compounds are produced from their non-volatile precursors. Because the sensory thresholds for volatiles are so low and metabolism nuances for the yeast strain so variable, it is crucial to determine the correct inoculum (its size, species and their ratio) or decide for spontaneous fermentation.
Ribolla Gialla plants are famous for their robust berries and their wines known for plain sensory profile. The production yields are high, but, unfortunately, the resulting beverage bouquet is rather poor. One possibility to increase the complexity of the wine is to perform spontaneous fermentations, but in many cases this is too risky, unpredictable and not reproducible. In our study, we offered an alternative solution in the form of sequential mixed-culture fermentations. Two yeast strains K. gamospora (CBS10400) and Z. kombuchaensis (CBS8849) contributed most to the aroma of the wine by adding an aroma of flowers, pear and honey. Importantly, these strains do not have a negative effect on the fermentation dynamics and perform well with the subsequently inoculated industrial S. cerevisiae strain.
Although the present research gives us useful and practical information, a possible drawback is the relatively small volume of laboratory and pilot fermentations. To solve this future industrial-scale fermentations are needed to efficiently qualify yeast industrial potential.
References
Bavčar D, Baša Česnik H, Čuš F, Košmerl T (2011) The influence of skin contact during alcoholic fermentation on the aroma composition of Ribolla Gialla and Malvasia Istriana Vitis vinifera (L.) grape wines. Int J Food Sci Technol 46:1801–1808. doi:10.1111/j.1365-2621.2011.02679.x
Beckner M, Ivey ML, Phister TG (2011) Microbial contamination of fuel ethanol fermentations. Lett Appl Microbiol 53:387–394. doi:10.1111/j.1472-765X.2011.03124.x
Bely M, Stoeckle P, Masneuf-Pomarède I, Dubourdieu D (2008) Impact of mixed Torulaspora delbrueckii-Saccharomyces cerevisiae culture on high-sugar fermentation. Int J Food Microbiol 122:312–320. doi:10.1016/j.ijfoodmicro.2007.12.023
Benda I (1982) Wine and brandy. In: Prescott SC, Dunn CG, Reed G (eds) Prescott & Dunn’s industrial microbiology, 4th edn. AVI Publishing Co., Westport, CN
Blomqvist J, Eberhard T, Schnürer J, Passoth V (2010) Fermentation characteristics of Dekkera bruxellensis strains. Appl Microbiol Biotechnol 87:1487–1497. doi:10.1007/s00253-010-2619-y
Boekhout T, Kurtzman CP, O’Donnell K, Smith MT (1994) Phylogeny of the yeast genera Hanseniaspora (anamorph Kloeckera), Dekkera (anamorph Brettanomyces), and Eeniella as inferred from partial 26S ribosomal DNA nucleotide sequences. Int J Syst Bacteriol 44:781–786
Comitini F, Gobbi M, Domizio P, Romani C, Lencioni L, Mannazzu I, Ciani M (2011) Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol 28:873–882. doi:10.1016/j.fm.2010.12.001
Conant GC, Wolfe KH (2007) Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Syst Biol. doi:10.1038/msb4100170
Coutinho R, Branco P, Monteiro M, Malfeito-Ferreira M, Albergaria H (2013) Saccharomyces cerevisiae and Dekkera bruxellensis interactions in alcoholic fermentations: growth and 4-ethylphenol production. MicroBiotec’13: Portuguese Congress of Microbiology and Biotechnology, Aveiro, Portugal, p 94
Dairou V, Sieffermann J-M (2002) A comparison of 14 jams characterized by conventional profile and a quick original method, the flash profile. J Food Sci 67:826–834. doi:10.1111/j.1365-2621.2002.tb10685.x
Dashko S, Zhou N, Compagno C, Piškur J (2014) Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Res 14:826–832. doi:10.1111/1567-1364.12161
De Deken RH (1966) The Crabtree effect: a regulatory system in yeast. J Gen Microbiol 44:149–156
Van Dijken JP, Scheffers WA (1986) Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol Lett 32:199–224. doi:10.1111/j.1574-6968.1986.tb01194.x
Domizio P, Romani C, Lencioni L, Comitini F, Gobbi M, Mannazzu I, Ciani M (2011) Outlining a future for non-Saccharomyces yeasts: selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisiae for grape juice fermentation. Int J Food Microbiol 147:170–180. doi:10.1016/j.ijfoodmicro.2011.03.020
Fleet GH (2003) Yeast interactions and wine flavour. Int J Food Microbiol 86:11–22
Galafassi S, Merico A, Pizza F, Hellborg L, Molinari F, Piškur J, Compagno C (2011) Dekkera/Brettanomyces yeasts for ethanol production from renewable sources under oxygen-limited and low-pH conditions. J Ind Microbiol Biotechnol 38:1079–1088. doi:10.1007/s10295-010-0885-4
Gobbi M, Comitini F, Domizio P, Romani C, Lencioni L, Mannazzu I, Ciani M (2013) Lachancea thermotolerans and Saccharomyces cerevisiae in simultaneous and sequential co-fermentation: a strategy to enhance acidity and improve the overall quality of wine. Food Microbiol 33:271–281. doi:10.1016/j.fm.2012.10.004
Gobbi M, De Vero L, Solieri L, Comitini F, Oro L, Giudici P, Ciani M (2014) Fermentative aptitude of non-Saccharomycescerevisiae wine yeasts for reduction in ethanol content in wine. Eur Food Res, Technol
Godoy L, Martínez C, Carrasco N, Ganga MA (2008) Purification and characterization of a p-coumarate decarboxylase and a vinylphenol reductase from Brettanomyces bruxellensis. Int J Food Microbiol 127:6–11. doi:10.1016/j.ijfoodmicro.2008.05.011
Guillaume C, Delobel P, Sablayrolles J-M, Blondin B (2007) Molecular basis of fructose utilization by the wine yeast Saccharomyces cerevisiae: a mutated HXT3allele enhances fructose fermentation. Appl Environ Microbiol 73:2432–2439. doi:10.1128/AEM.02269-06
Hagman A, Säll T, Compagno C, Piskur J (2013) Yeast “make-accumulate-consume” life strategy evolved as a multi-step process that predates the whole genome duplication. PLoS One 8:e68734. doi:10.1371/journal.pone.0068734
Van Hoek P, Van Dijken JP, Pronk JT (1998) Effect of specific growth rate on fermentative capacity of baker’s yeast. Appl Environ Microbiol 64:4226–4233
Jolly NP, Varela C, Pretorius IS (2014) Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res 14:215–237. doi:10.1111/1567-1364.12111
Jussier D, Dubé Morneau A, Mira de Orduña R (2006) Effect of simultaneous inoculation with yeast and bacteria on fermentation kinetics and key wine parameters of cool-climate Chardonnay. Appl Environ Microbiol 72:221–227. doi:10.1128/AEM.72.1.221-227.2006
Kim D-H, Hong Y-A, Park H-D (2008) Co-fermentation of grape must by Issatchenkia orientalis and Saccharomyces cerevisiae reduces the malic acid content in wine. Biotechnol Lett 30:1633–1638. doi:10.1007/s10529-008-9726-1
Kurtzman CP, Robnett CJ (1997) Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol 35:1216–1223
Kurtzman CP, Robnett CJ (2003) Phylogenetic relationships among yeasts of the “Saccharomyces complex” determined from multigene sequence analyses. FEMS Yeast Res 3:417–432
Lambrechts MG, Pretorius IS (2000) Yeast and its importance to wine aroma—a review. South Afr J Enol Vitic, South Afr
Lawless HTHH (2010) Sensory evaluation of food—principles and practices. Springer Science and Business Media, New York
Luca Riccardo Formenti AN (2014) Challenges in industrial fermentation technology research. Biotechnol J. doi:10.1002/biot.201300236
Øyvind Langsrud TN (1998) A unified framework for significance testing in fractional factorials. Comput Stat Amp Data Anal. doi:10.1016/S0167-9473(98)90151-7
Pao SS, Paulsen IT, Saier MH (1998) Major Facilitator Superfamily. Microbiol Mol Biol Rev 62:1–34
Piskur J, Langkjaer RB (2004) Yeast genome sequencing: the power of comparative genomics. Mol Microbiol 53:381–389. doi:10.1111/j.1365-2958.2004.04182.x
Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C (2006) How did Saccharomyces evolve to become a good brewer? Trends Genet TIG 22:183–186. doi:10.1016/j.tig.2006.02.002
Rainieri S, Pretorius IS (2000) Selection and improvement of wine yeasts. Ann Microbiol 50:15–31
Rantsiou K, Dolci P, Giacosa S, Torchio F, Tofalo R, Torriani S, Suzzi G, Rolle L, Cocolin L (2012) Candida zemplinina can reduce acetic acid produced by Saccharomyces cerevisiae in sweet wine fermentations. Appl Environ Microbiol 78:1987–1994. doi:10.1128/AEM.06768-11
Ravasio D, Walther A, Trost K, Vrhovsek U, Wendland J (2014) An indirect assay for volatile compound production in yeast strains. Sci Rep 4:3707. doi:10.1038/srep03707
Romano A, Perello MC, Lonvaud-Funel A, Sicard G, de Revel G (2009) Sensory and analytical re-evaluation of “Brett character”. Food Chem 114:15–19. doi:10.1016/j.foodchem.2008.09.006
Rozpędowska E, Hellborg L, Ishchuk OP, Orhan F, Galafassi S, Merico A, Woolfit M, Compagno C, Piskur J (2011) Parallel evolution of the make-accumulate-consume strategy in Saccharomyces and Dekkera yeasts. Nat Commun 2:302. doi:10.1038/ncomms1305
Schifferdecker AJ, Dashko S, Ishchuk OP, Piškur J (2014) The wine and beer yeast Dekkera bruxellensis: the wine and beer yeast Dekkera bruxellensis. Yeast 31:323–332. doi:10.1002/yea.3023
Semchyshyn HM, Abrat OB, Miedzobrodzki J, Inoue Y, Lushchak VI (2011) Acetate but not propionate induces oxidative stress in bakers’ yeast Saccharomyces cerevisiae. Redox Rep Commun Free Radic Res 16:15–23. doi:10.1179/174329211X12968219310954
Sousa-Dias SGT (1996) Kinetics and regulation of fructose and glucose transport systems are responsible for fructophily in Zygosaccharomyces bailii. Microbiol-Sgm 142:1733–1738. doi:10.1099/13500872-142-7-1733
Stone H, Sidel JL (1998) Quantitative descriptive analysis: developments, applications and the future. Food Technol, USA
Suárez R, Suárez-Lepe JA, Morata A, Calderón F (2007) The production of ethylphenols in wine by yeasts of the genera Brettanomyces and Dekkera: a review. Food Chem 102:10–21. doi:10.1016/j.foodchem.2006.03.030
Tiukova IA, Petterson ME, Tellgren-Roth C, Bunikis I, Eberhard T, Pettersson OV, Passoth V (2013) Transcriptome of the alternative ethanol production strain Dekkera bruxellensis CBS 11270 in sugar limited, low oxygen cultivation. PLoS One 8:e58455. doi:10.1371/journal.pone.0058455
Toro ME, Vazquez F (2002) Fermentation behaviour of controlled mixed and sequential cultures of Candida cantarellii and Saccharomyces cerevisiae wine yeasts. World J Microbiol Biotechnol 18:351–358. doi:10.1023/A:1015242818473
Verduyn C, Postma E, Scheffers WA, Van Dijken JP (1992) Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast Chichester Engl 8:501–517. doi:10.1002/yea.320080703
Wedzicha BL (1984) Chemistry of sulphur dioxide in foods. Elsevier Applied Science, London, New York
Wehrens R, Weingart G, Mattivi F (2014) metaMS: an open-source pipeline for GC–MS-based untargeted metabolomics. J Chromatogr B 966:109–116. doi:10.1016/j.jchromb.2014.02.051
White T, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Shinsky J, White T (eds) PCR Protoc. Academic Press, Guide Methods Appl, pp 315–322
Wolfe K (2004) Evolutionary genomics: yeasts accelerate beyond BLAST. Curr Biol CB 14:R392–R394. doi:10.1016/j.cub.2004.05.015
Wolfe KH, Shields DC (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708–713. doi:10.1038/42711
Yamaoka C, Kurita O, Kubo T (2014) Improved ethanol tolerance of Saccharomyces cerevisiae in mixed cultures with Kluyveromyces lactis on high-sugar fermentation. Microbiol Res 169:907–914. doi:10.1016/j.micres.2014.04.007
Green SR, Gray PP (1950) A differential procedure applicable to bacteriological investigation in brewing. Wallerstein Lab. Commun 13:357
Gamero Lluna A, de Jong C (2013) Novel yeasts, novel flavours. New Food Mag. 16(3)26–28
Acknowledgments
This work was supported by the Slovenian Research Agency (project number J4-4300) and by the EU ITN “Cornucopia” project (FP7, grant agreement GA264717), Creative Core programme (AHA-MOMENT) contract no. 3330-13-500031, co-supported by RS-MIZS and European Regional Development Fund Research. We would like to thank Justin Fay for providing useful advice and corrections during the writing process. We greatly appreciate the access to the yeast strains provided by CBS-KNAW Fungal Biodiversity Centre (http://www.cbs.knaw.nl/). We are especially grateful to the late Jure Piškur, who was our main inspiration and project leader and who is greatly missed.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10295_2015_1620_MOESM1_ESM.pdf
Supplementary material 1 (PDF 124 kb). Weight loss during the fermentation of laboratory-scale fermentations performed with D. anomala (CBS77), D. bruxellensis (CBS2796), K. gamospora (CBS10400), T. microellipsoides (CBS6641), Z. kombuchaensis (CBS8849). The steeper is the curve the higher is fermentation dynamic of the sample
10295_2015_1620_MOESM2_ESM.pdf
Supplementary material 2 (PDF 349 kb). PCA plot with most influential loadings of six yeast species based on the major volatiles formed during the fermentation process. Fermentation samples produced with DA - D. anomala (CBS77), DB - D. bruxellensis (CBS2796), KG - K. gamospora (CBS10400), TM - T. microellipsoides (CBS6641), ZK - Z. kombuchaensis (CBS8849)
10295_2015_1620_MOESM3_ESM.pdf
Supplementary material 3 (PDF 279 kb). Results of sensory evaluation of young wines using hedonic scale method. 1 – least preferred sample, 10 – most preferred one. (a) evaluation of “smell” attribute in six samples of the young wines. (b) evaluation of “color” attribute in six samples of the young wines. Fermentation samples produced with DA - D. anomala (CBS77), DB - D. bruxellensis (CBS2796), KG - K. gamospora (CBS10400), TM - T. microellipsoides (CBS6641), ZK - Z. kombuchaensis (CBS8849)
10295_2015_1620_MOESM4_ESM.pdf
Supplementary material 4 (PDF 348 kb). PCA plot demonstrating patterns of variations of sensory attributes. (a) Z. kombuchaensis (CBS8849) and K. gamospora(CBS10400) are mostly impacting aroma attribute (in red), in less extent attributes of color (in blue) and taste (in green). (b) wines produced with K. gamospora are more distinguishable from S. cerevisiae Lalvin T73 produced wines in comparison to Z. kombuchaensis samples
10295_2015_1620_MOESM5_ESM.pdf
Supplementary material 5 (PDF 104 kb). Levels of sugars and ethanol in the pilot scale fermentation samples fermented with K. gamospora (CBS10400), Z. kombuchaensis (CBS8849) and S. cerevisiae Lalvin T73 after 350 h of fermentation. Statistical groups determined using LSD test
10295_2015_1620_MOESM6_ESM.pdf
Supplementary material 6 (PDF 376 kb). Comparative physiology of the control strain, S. cerevisiae Lalvin T73 andK. gamospora (CBS10400), Z. kombuchaensis (CBS8849).Yields (ethanol, acetate, biomass, glycerol and pyruvate) were calculated during the exponential phase as a function of glucose consumed. Corresponding consumption and production rates were calculated during the same time intervals on minimal media supplied with 2% glucose in batch fermentations. a Maximum specific growth rate. b Yield coefficients per gram of glucose consumed (g−1); Yse, yield of ethanol; Ysx, yield of biomass; Ysp, yield of pyruvate; Ysac, yield of acetate; Ysg, yield of glycerol. c Specific consumption rate per gram of biomass per hour (mmol g−1 h−1); qGlucose, glucose consumption rate; qO2, oxygen consumption rate. d Specific production rates per hour per gram of biomass (mmol g−1 h−1);qEthanol, ethanol production rate; qCO2, carbon dioxide consumption rate. e Respiratory Quotient ; ratio of carbon dioxide production rate as per oxygen consumption rate
10295_2015_1620_MOESM8_ESM.pdf
Supplementary material 8 (PDF 67 kb). Ethanol, glucose, and fructose measurements of the samples taken after 60 h, 150 h, and 300 h. Data was processed using one-way ANOVA (n.s.: not significant; *: P<0.05 (*); **: P<0.01; ***: P<0.001)
10295_2015_1620_MOESM9_ESM.pdf
Supplementary material 9 (PDF 169 kb). (a) Concentrations of volatiles expressed as 2-octanol using chromatographic peak area ratios at the end of laboratory scale fermentations. (b) Concentrations of volatiles expressed as 2-octanol using chromatographic peak area ratios at the end of pilot scale fermentations
10295_2015_1620_MOESM11_ESM.pdf
Supplementary material 11 (PDF 382 kb). Raw data of sensory attributes with statistical data attached. Data was processed using one-way ANOVA (n.s.: not significant; *: P<0.05 (*); **: P<0.01; ***: P<0.001). Groups determined using LSD test
10295_2015_1620_MOESM12_ESM.pdf
Supplementary material 12 (PDF 63 kb). Raw data from HPLC analysis of basic metabolites from pilot scale fermentation samples fermented with K. gamospora (CBS10400), Z. kombuchaensis (CBS8849) and S. cerevisiae Lalvin T73. Statistical analysis attached, groups determined using LSD test
Rights and permissions
About this article
Cite this article
Dashko, S., Zhou, N., Tinta, T. et al. Use of non-conventional yeast improves the wine aroma profile of Ribolla Gialla. J Ind Microbiol Biotechnol 42, 997–1010 (2015). https://doi.org/10.1007/s10295-015-1620-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-015-1620-y