Abstract
The aim of this study was to validate a novel medical virtual reality (VR) platform used for medical image segmentation and contouring in radiation oncology and 3D anatomical modeling and simulation for planning medical interventions, including surgery. The first step of the validation was to verify quantitatively and qualitatively that the VR platform can produce substantially equivalent 3D anatomical models, image contours, and measurements to those generated with existing commercial platforms. To achieve this, a total of eight image sets and 18 structures were segmented using both VR and reference commercial platforms. The image sets were chosen to cover a broad range of scanner manufacturers, modalities, and voxel dimensions. The second step consisted of evaluating whether the VR platform could provide efficiency improvements for target delineation in radiation oncology planning. To assess this, the image sets for five pediatric patients with resected standard-risk medulloblastoma were used to contour target volumes in support of treatment planning of craniospinal irradiation, requiring complete inclusion of the entire cerebral–spinal volume. Structures generated in the VR and the commercial platforms were found to have a high degree of similarity, with dice similarity coefficient ranging from 0.963 to 0.985 for high-resolution images and 0.920 to 0.990 for lower resolution images. Volume, cross-sectional area, and length measurements were also found to be in agreement with reference values derived from a commercial system, with length measurements having a maximum difference of 0.22 mm, angle measurements having a maximum difference of 0.04°, and cross-sectional area measurements having a maximum difference of 0.16 mm2. The VR platform was also found to yield significant efficiency improvements, reducing the time required to delineate complex cranial and spinal target volumes by an average of 50% or 29 min.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Background
Accurate and specific organ delineation on patient medical images constitutes a vital element of many medical specialties. Segmentations, contours, and measurements on medical images may be used for communication, diagnosis, evaluating disease progression, planning subsequent interventions, or optimizing treatments, among others [1].
In the absence of automation driven by deep learning, or some other viable alternative, many delineation platforms require users to outline anatomical structures on medical images “slice-by-slice,” which is both consuming of time and attention. While semi-automated and fully automated methods exist [1,2,3], they are not guaranteed to always achieve the desired or required result in the clinical setting. Even for anatomical structures routinely segmented for planning purposes, image segmentations or contours may require manual input and adjustment.
In this work, we report on the use of a novel medical image delineation platform based on modern consumer virtual reality (VR) technology. Theoretically, the use of VR provides the potential to perform volumetric contouring as well as the ability to contour in planes beyond the three conventional cardinal planes. The physical extent, alignment, separation, and other dimensional properties become more perceptible as the user maneuvers through the virtual space in the immersive environment [4]. One situation in which this could be particularly advantageous is for disease sites where the definition of the delineated volume is based on the 3D spread of disease along anatomical features with intricate topology. It can be challenging and time-consuming to precisely contour and measure along a curved structure that may not lie neatly in any of the traditional axial, sagittal, or coronal planes [5].
The VR platform under consideration here was initially designed and developed at our institution and recently commercialized under the name Elucis™ (Realize Medical Inc., Ottawa, Canada) [6]. The platform was recently implemented clinically at our institution in the Department of Radiology, Radiation Oncology, and Medical Physics for use in several research ethics-approved trials. The clinical evaluation of Elucis in this work involves (1) a technical comparison with reference commercial desktop systems for medical image segmentation and contouring and (2) an assessment of the overall general usability in the clinical setting as measured by the time required to achieve the desired clinical result.
Prior attempts at using VR to achieve medical image contouring in support of radiation therapy treatment planning include VRContour [7, 8] and DICOM VR [5, 9]; however, there is no indication that any of the creators made attempts to iteratively improve their user interface to suit the needs of end-users and get clinical approval from regulatory agencies. A usability follow-up study on the usability of the Elucis software involving 18 clinicians volunteers will be presented in a separate study (in preparation).
Elucis has previously been evaluated for its utility in, specifically, mandible segmentation using a public dataset of ten CT scans and five study participants [10]. That investigation found Elucis was slightly superior to the artificial intelligence and desktop screen-based segmentation systems used in the comparison in terms of speed and accuracy (as demonstrated by metrics and judged by expert reviewers). A Likert questionnaire completed by the participants also revealed that Elucis was easier to use for preparing a mandible model with precision and for appreciating unique anatomical characteristics. This initial evaluation, however, was limited to boney anatomy segmented from CT scans. As such, in this work, we expand on the evaluation of Elucis by assessing its utility in modeling several bone and soft-tissue structures from CT and MR images. One could postulate that such a system would also require a technical assessment of the associated VR tracking system positional accuracy; however, errors in positional tracking are of little consequence as the system is entirely self-contained — i.e., being a VR application (as opposed to an augmented reality system utilizing auto-registration), it does not rely on relating positions in the virtual environment to locations or objects in the real world. That said, tracking errors could, at least theoretically, disrupt the sense of immersion in the VR environment, so ensuring these errors are minimized is of general importance. Incidentally, several investigations into the accuracy of tracking motion have revealed that the tracking systems compatible with Elucis have small tracking errors. In one example, the Oculus VR system (Meta, Menlo Park, California) was used to measure the dimensions of various scanned objects. Linear and angular measurement errors were estimated to be 0.5 mm and 0.7°, respectively [11]. In another study, the Vive Pro VR system (HTC, Taoyuan, Taiwan) was combined with the Polaris Spectra optical tracking system (NDI, Waterloo, Ontario, Canada) to perform measurements on physical objects. The reported positional accuracy and precision were 0.48 mm and 0.23 mm, respectively, while the rotational accuracy and precision results were, respectively, 0.64° and 0.05°. These reported discrepancies are inconsequential to the sense of immersion one experiences in VR and more importantly, as will be shown, do not contribute to inaccuracies in 3D models created in VR.
While tracking errors associated with consumer VR system are well documented, to our knowledge, there are no publications reporting on the accuracy and feasibility of using clinically approved VR products for manual and semi-automated medical image processing. In this study, we report such results. The first part of the study focuses on the ability of the VR image processing platform considered here to produce image segments and contours that are substantially equivalent to those generated with popular commercial alternatives, namely, Mimics (Materialise, Leuven, Belgium) and Monaco (Elekta, Stockholm, Sweden). The second part of this work examines the feasibility and benefits, if any, of using VR for medical image processing. In the latter initiative, the time required to make contours of complex cranial–spinal target volumes from standard CT simulation images was measured for both the VR platform and the reference system.
Methods
Accuracy of Image Segmentation and Contouring: Virtual Reality vs Reference Desktop Platform
A variety of anatomical and geometrical structures was segmented from CT and MR images using the Elucis™ medical VR platform (Realize Medical Inc., Ottawa, Canada). Image sets were chosen to cover a range of scanner manufacturers, modalities, and voxel dimensions. The resulting three-dimensional (3D) structures intended for 3D surgical planning were compared with the commercial desktop platform Mimics™ (Materialise, Leuven, Belgium), while those intended for radiation oncology treatment planning were compared with Monaco™ (Elekta, Stockholm, Sweden). All images used in this study were anonymized in accordance with the research ethics board-approved protocol governing the conduct of this investigation.
With VR, the ways that the medical structures are rendered are somewhat critical, which affect how participants understand the structures. In Elucis, structures are stored internally as 3D signed distance fields (SDF). The distance fields are rendered in two ways: two-dimensional (2D), or cross-sectional, visualization with or without overlay on 2D image cross-sections is presented by creating the classic outline and fill representations using a custom render shader that samples the SDF in 3D space and draws a solid or transparent color accordingly.
Three-dimensional visualization of the structures is achieved by applying the marching cubes algorithm to construct high-resolution 3D meshes that define the surface (zero-set boundary) described by the SDF.
Exported structures were compared quantitatively and qualitatively for geometric equivalence. In the absence of a ground truth for these structures, those exported from the conventional desktop platforms served as the reference for the Elucis VR platform. A total of 6 image sets and 8 structures were segmented for comparison with Mimics by an experienced medical physicist with over 10 years of clinical experience. An additional two image sets and ten structures were segmented for comparison with Monaco by another experienced medical physicist. All segmented or contoured structures from all platforms were reviewed for accuracy by an experienced (> 10 years) radiologist reviewer in the axial, sagittal, and coronal images planes.
For structures created using semi-automated methods involving image thresholds, the identical image value threshold ranges were used in both platforms. Structures in Elucis were performed with the “add” function combined with HU threshold selection. Structures in Mimics were processed with the “Convert to Part” function to generate a 3D mesh. Structures in Monaco were generated with the automated thresholding tool to create 2D contours on each relevant slice, interpolating where appropriate to optimize efficiency while maintaining accuracy. In most cases, the user applied minor edits to achieve the most accurate results. Structures from Elucis and Mimics were exported in STL format to facilitate a comparison in 3D Slicer. For comparisons in Monaco, image segmentations in Elucis were exported in DICOM-RT format. The image set properties and corresponding geometries are described in Table 1.
DICOM volume information is transferred into Elucis by reading in the voxel information and storing it in a 3D texture array. This 3D array is attached to a corresponding virtual object with dimensions, position, rotation, and size as described in DICOM header. 2D contours for exported DICOM-RT files are generated by creating a virtual plane at each position as defined by the DICOM image planes and then sampling the 3D signed distance fields on a high-resolution grid. The marching squares algorithm is then applied to the resulting distance-defining pixels to produce a set of line segments that define the contour points. Since the methods for structure generation, storage, and export rely on the implicit surface of the 3D signed distance field, surface definition precision is well below a voxel width in size.
Quantitative Comparisons
Validation of Sørensen–Dice Similarity Coefficient (DSC) Calculation Accuracy
Quantitative analysis of 3D structures was performed by comparing structure files and image sets exported from each system in the open-source image computing platform 3D Slicer (vs 4.10.2) [12].The Sørensen–Dice similarity coefficient (DSC) [13] was used to quantify the similarity between structures generated in VR and reference commercial platforms. The Hausdorff distance metrics was also used for the patient structures and some irregular structures. The Slicer DCS algorithm was first validated using the segmentation of a 6 × 6 × 6 cm3 cube structure. Duplicates of the cube were created with a positive lateral shift in one direction of 3 mm, 10 mm, and 20 mm. Using the shifts and known volumes of the cubes, the true DSC can be calculated analytically and compared to the DSC values reported by Slicer.
DCS Scores for Segmented Structures and Measurement Accuracy
A collection of anatomical and 3D simple geometries were segmented from eight medical images, covering a variety of image types and slice thicknesses (Table 1). From these same eight image sets, length, angle, and cross-sectional area measurements were also performed in both Elucis and Mimics. Measurements were conducted on a mix of cross-sectional image views and on 3D models generated from image segmentations.
Qualitative Comparisons
Visual comparison of structures was performed by importing structure files generated in the VR platform into the corresponding reference commercial desktop platform: the segmented or contoured structures from all platforms were reviewed for accuracy by an expert radiologist reviewer in the axial, sagittal, and coronal images planes.
Efficiency of Complex Volume Segmentation for Radiotherapy Treatment Planning: Virtual Reality vs a Reference Desktop Platform
In the second part of this investigation, we measured the time efficiency of VR-based segmentation and contouring relative to the reference commercial platform Monaco. Five pediatric patients with resected standard risk medulloblastoma were retrospectively selected for this ethics-approved study.
For each patient, a computed tomography (CT) image set was acquired using 2 mm slice thickness to delineate clinical target volumes (CTV) in support of treatment planning of craniospinal irradiation. Image sets were imported into both the Elucis VR platform and the reference commercial treatment planning system from Elekta (Monaco). Cranial and spinal clinical target volumes (CTVs) were contoured as per the European Society for Pediatric Oncology (SIOPE) consensus guidelines [14] for medulloblastomas as shown in Table 2. Semi-automatic contouring tools, including interpolation tools, were leveraged as much as possible in both systems.
Each contour was generated by two clinicians: a junior radiation oncology resident with 3 h of training time in Elucis and Monaco and a medical physicist with 19 years of experience with contouring software platforms, including over 10 years with the reference Monaco treatment planning system. The time to contour target structures (CTV cranial, CTV spinal, total time) was compared between both platforms.
This approach helped minimize potential bias due to disparity in familiarity, as users at our institution are intimately familiar with reference commercial platform, while the VR delineation platform has only become available very recently.
Results
Accuracy of Image Segmentation and Contouring: Virtual Reality vs Reference Desktop Platform
Quantitative Comparisons
Validation of DSC Calculation Accuracy
Table 3 summarizes the results of DSC values reported by the 3D Slicer software system for a 6 × 6 × 6 cm3 cube structure shifted by known distances from the reference cube structure. These values differ from expected results, calculated analytically, by a maximum of eight parts in 1000, or 0.8%; however, we note the percentages error is the largest for small DSC values. Minor differences between these analytically calculated values and the results obtained with 3D Slicer are likely attributed to uncertainties when shifting the contours since the contour points are anchored to the nearest voxel center. The voxel size of the image used to contour the cube is 0.6 mm. It implies an uncertainty of 0.3 mm in the shift or 0.5% to 0.8% in the DSC values depending the size of the shift.
DCS Scores for Segmented Structures and Measurement Accuracy
A mix of anatomical and simple geometric structures generated in the Elucis VR platform and reference commercial platforms were found to have a high degree of similarity, with DSC values ranging from 0.963 to 0.985 for high-resolution images and 0.920 to 0.990 for lower resolution images. Lower DSC scores are expected for structures defined on image sets reconstructed with large pixel size, especially as the structure size decreases. The sensitivity of volume to pixel size for some structures was estimated by calculating the change in volume when we shrink or expand the structure by half a pixel in all directions.
Measurements of length and angle were found to be comparable between Elucis and reference commercial platforms, with length measurements having a maximum difference of 0.22 mm, angle measurements having a maximum difference of 0.04°, and cross-sectional area measurements having a maximum difference of 0.16 mm2. Specific values for DSC scores and Hausdorff distance values are shown in Table 4 along with linear and angular measurements. Specific values for DSC scores and sensitivity to voxel size for a subset of the geometric shapes are shown in Table 5.
Qualitative Comparisons
A visual comparison of structures created in Elucis and reference commercial platforms is presented in Figs. 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, and 15. Length and/or angle measurements are also displayed throughout, depending on the image under consideration, as described in the corresponding image captions and accompanying summary of results.
Visual comparison of primitive structures created using Mimics (cyan) and Elucis (red). See Fig. 2 for a zoom
a A comparison between Mimics contours (red) and Elucis contours (green) of the cone structure displayed in the coronal view b Of note is the placement of the superior-most (z-direction) boundary in the center of the voxels for the Mimics structure, while the Elucis structure correctly encompasses the desired voxels
Length measurement of the 3D blood pool structure from Fig. 10 in a Mimics and b Elucis
Length measurements of the brain tumor from Fig. 10 in a Mimics and b Elucis
Segmentations of Primitive Structures (Cone, Cylinder, and Cube) from the TG132 Computerized Geometric Phantom
A comparison of primitive structures (i.e., a cone, sphere, cylinder, and cube) segmented using Elucis (cyan) and Materialise Mimics (red) on the TG132 geometric phantom is shown in Fig. 1. This comparison was conducted in Mimics by imported structures created in Elucis.
Figure 2 shows a close-up of the cone structure in the coronal view (left), with a further close-up of the superior boundary of the cone (right). In this figure, the model of the cone made using Elucis is shown in green, and that made with Mimics in red. These structures were created in each platform using the same image value threshold range applied to the image prior to segmentation of the desired image region. A gap between the two resulting structures can clearly be observed along the superior boundary of the cone, highlighting a stark difference between the resulting structures from each platform. The structure created in Elucis tightly conforms to the superior edge of the cone on all slices, while the superior limit of the structure created with Mimics is offset from this superior edge of the cone by one half of a voxel width (slice thickness).
Figure 3 shows line and angle measurements conducted in Elucis and Mimics for the same image containing the primitive structures. Here, measurement values agree between the two platforms (Table 4), with residual differences attributed the user’s ability to discern boundaries and accurately place a point.
Kidney Segmentation from a CT with Contrast (2.5 mm Slice Thickness)
A visual comparison of kidney structures segmented in both Elucis and Mimics from a CT scan can be seen in Fig. 4. Segmented structures from both platforms tightly conform to the radiological boundaries of the kidney, while 3D views of the respective structures (bottom right of figure) are visually indistinguishable. Figure 5 shows length measurements in the sagittal view of the kidney from both platforms yielding the same results within 0.2 mm.
Mandible Segmentation from a CT of the Skull (0.625 mm Slice Thickness)
Figure 6 shows segmentations from Elucis (red) and Mimics (cyan) of the right side of a fractured mandible, created from a CT of the skull with 0.625-mm-thick slices. As with the kidney example shown in Fig. 4, the segmented structures from both platforms are visually identical. A length measurement of the fractured mandible was performed using a 3D view of the structure in Elucis and Mimics, as displayed in Fig. 7. The measured lengths are, incidentally, within 0.1 mm.
Segmentations of Lungs and Airways from a CT of the Chest (1 mm Slice Thickness)
The lungs and airways were created from a chest CT with 1-mm-thick slices. A visual comparison of the lungs and airways can be seen in Fig. 8, which shows the result from Elucis (red) and Mimics (yellow). An angle measurement of the carina was performed using the coronal image view using both platforms and can be seen in Fig. 9.
Cardiac Blood Pool Segmentations from a CTA of an Adult Heart (0.3 mm Slice Thickness)
Models of the left-sided cardiac blood pool derived using Elucis and Mimics from a computed tomography angiogram (CTA) of an adult heart are shown in Fig. 10. The structures are visually indistinguishable apart from where the user chose to end the inclusion of pulmonary veins and arteries, although the model boundaries were made identical in the final comparison. Length measurements, shown in Fig. 11, were conducted using the 3D structure view in the respective platforms and yielded the same results within 0.1 mm. Cross-sectional area measurements conducted in both platforms of the base of the aorta as shown in the axial image view are shown in Fig. 12. Results of cross-sectional area are with 0.1 mm2.
Brain Tumor Segmentation for a T1 MRI
Figure 13 shows segmentations of a brain tumor on a T1-weighted MRI sequence using both Elucis (red) and the reference commercial platform Mimics (yellow). As with the segmentations conducted on CT images, the resulting structures are visibly indistinguishable from each other. Length measurements of the tumor captured by both platforms in the axial image view are shown in Fig. 14 and agree within less than 0.1 mm.
Contours of Cylindrical Rod in a Geometric CT Phantom (1 mm and 3 mm Slice Thickness)
Lastly, Fig. 15 shows a visual comparison of cylindrical rod contours generated with Elucis (blue) and the reference commercial platform Monaco (red) from a CT of a geometric phantom. Here, contours were generated for reconstructed slice thicknesses of 1 and 3 mm. The contours from both platforms are visually indistinguishable.
Efficiency of Complex Volume Segmentation for Radiotherapy Treatment Planning: Virtual Reality vs a Reference Desktop Platform
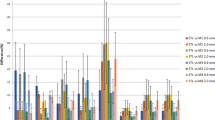
Contours in support of radiotherapy planning of pediatric patients indicated for craniospinal irradiation are shown in Fig. 16. As a test of general utility, all organs at risk for radiation-induced injury were also contoured in the Elucis VR platform for planning purposes, including all bony structures, eyes, and optic structures (optic nerves and chiasm), brainstem, parotid glands, thyroid, heart, lungs, liver, spleen, kidneys, bowel, and bladder (Fig. 16a). The target volumes generated with Elucis for the five patients are shown in Fig. 16b. VR-based contouring significantly reduced the time required to generate (contour) these complex cranial and spinal target volumes by an average of 41% (35 min) for a junior radiation oncology resident new to both platforms and 58% (22 min) for an experienced medical physicist. The average contouring time for the experienced user was 59% and 73% faster for the standard and the VR delineation platform, respectively. Detailed results for each structure can be seen in Fig. 17. The difference between platforms was determined to be highly statistically significant (p < 0.001).
Discussion
Linear and angular measurements performed in the VR delineation platform are substantially equivalent to those performed in the standard delineation platform, with differences not exceeding 0.22 mm and 0.04°, respectively, which is smaller than half of the smallest image set pixel size used in this study. As such these small differences could be attributed to the users’ ability to resolved ambiguity in the image (e.g., an image boundary) or to the ability to accurately recreate the placement of a point in space. Despite the good agreement observed between segmentation and contours between the virtual reality system and the reference commercial desktop platforms, we acknowledge this study is limited by the number of participants.
The level of agreement between structures created in the respective platforms was assessed using DSC and average Hausdorff distance scores. Using DSC scores to evaluate if two structures are substantially equivalent requires some consideration. The lowest DSC score observed in this study is 0.92 (large bone rod cylinder), which implies that the non-overlapping regions represent about 8% of the average structure volume. To determine if this is acceptable, one should consider the sensitivity of the structure volume to pixel size by estimating the change in volume when we shrink the structure by half a pixel in all directions. Half a pixel is a reasonable choice because different segmentation algorithms based on pixel value threshold may delineate contour boundaries at different locations within voxels. For example, Fig. 2 shows that the reference commercial platform assigned the structure boundary to the center of the last voxel, while the VR delineation platform encompasses the full extent of the voxel. The impact on a DSC score is not negligible when the structure is small and the slice thickness is large (in this example, it was 3 mm). In the case of the large bone rod structure, a result of 0.92 (8% of structure volume) is acceptable since a change of half a pixel in each direction creates a difference of 17% in volume. For structures like these, the Hausdorff distance metric, which provides a measure of how far apart two points meant to be co-located are from each other, serves as a more meaningful metric for quantifying the difference between two structures. In this study, the average calculated Hausdorff distances were all below 0.7 mm, while the 95th percentile was below 2.5 mm, indicating excellent geometric equivalence between structures segmented with Elucis and the reference desktop platform from Materialise. It should be noted, however, that residual differences could be attributed to how the respective platforms determine boundaries for a given image value threshold. In particular, an artifact of the “convert to part” functionality in Mimics leads to an inaccurate boundary that straddles the mid-point of a voxel within a structure rather than its outer edge. This is clearly demonstrated using the cone structure that was segmented from the TG132 phantom in the respective platforms, as shown in Fig. 1. Incidentally, the segmentations of primitive structures in this phantom, which had 3-mm-thick slices, resulted in the worst DSC and Hausdorff distance scores.
The second part of the study aimed to determine if using the VR delineation platform is more efficient than using a reference commercial desktop platform for a specific use case, namely the highly specific contouring of craniospinal target volumes in pediatric patient simulation CT. One limitation of this study is that the contours were performed on a small number of cases (5) by only two users. It can be challenging to precisely contour a curved structure that may not lie neatly in any of the traditional axial, sagittal, or coronal planes. This especially applies to the present challenge of defining spinal canal volume which, for these patients, must include the bilateral foramen through which the nerve roots traverse. As such, this study required users to take care to ensure the entire craniospinal volume containing the cerebrospinal fluid was included to facilitate the prescription of radiation dose to those areas: The cranial structure must include the cribriform plate, the pituitary fossa and dural cuffs of cranial nerves as they pass through the skull base foramina, while the spinal structure must include the bilateral nerve roots and continue through the sacral nerve roots to the termination of the dural sac. Using the Elucis VR platform for this task was about twice as fast for both the experienced and the non-experienced user. Both users attributed this to the freedom to navigate the image series in a more natural, immersive environment that makes it easier to access available image information. With sufficient training and experience, a user should become adept at generating the same structures in VR and in reference commercial desktop platforms since they both display the same underlying information; however, any reduction in cognitive load and improvements in perceptibility when using the VR platform should ultimately translate, in principle, to improvements in efficiency. Efficiency improvements associated with using the Elucis VR system over the conventional desktop platform may also be explained by differences in the segmentation and contouring tools offered in the respective platforms. This study did not differentiate between tools and is also limited by the fact that it is based on only two users with different level of experience.
We note the experience of the user and familiarity with segmentation tools made a significant difference. The average contouring time for the experienced user was 59% and 73% faster for the standard and the VR delineation platform, respectively. For the non-experienced user, the improvements in contouring efficiency were similar for both cranial and spinal structures and were mostly derived from easy access to non-standard contouring planes (cross-sectional views) in the VR platform. For the experienced user, the improvements in contouring efficiency were more important for the spinal structures because the standard delineation platform required the user to manually contour the spinal structures on each transverse slice to fully include nerve roots. The VR delineation platform offered a way to quickly generate the spinal cord structure plus some expansion on a single plane, while the editing tools made it possible to quickly smooth the nerve roots exiting the bone. For the experienced user, the improvements in contouring efficiency for the brain structures were not as important because no single plane offered an easy path to quickly access all the structures and the standard delineation platform included semi-automated thresholding tools to automatically contour the interface between the brain tissue and the cortical bone. In Elucis, the thresholding tools were more generic, but using the platform was still faster because generating and cleaning the contours were faster in 3D compared with taking a “slice-by-slice” approach.
Conclusion
As part of its clinical implementation, the medical virtual reality platform Elucis was evaluated for its ability to generate 3D anatomical models in support of surgical planning, medical 3D printing, and radiotherapy treatment planning accurately and efficiently. With respect to accuracy of medical image segmentation and contouring, comparisons with reference commercial desktop platforms via DSC and Hausdorff distance metrics demonstrated that image segmentations conducted in Elucis are substantially equivalent those obtained by standard approaches. The lowest observed DSC score from all comparisons was 0.92 which, in that particular case, corresponded to error of less than half of a voxel width. Otherwise, DSC scores were above 0.95. Likewise, average Hausdorff distances were at or below 0.7 mm for all structures, with the largest values attributed, at least in part, to artifacts in the way the reference desktop platform converts segmentations to structures. These DSC and Hausdorff distance observations are corroborated by visual assessment of the segmentations and their associated 3D models, where those produced using Elucis were found to be visually indistinguishable from the corresponding references.
The Elucis VR platform was also found to yield accurate measurements. Line, angle, and cross-sectional area measurements were found to agree with reference values within 0.3 mm, 0.04°, and 0.16 mm2, respectively.
Lastly, and perhaps most importantly, the general clinical utility of taking a VR-based approach to image contouring was established by the efficiency of this activity relative to a reference commercial desktop platform. Using VR was found to reduce the time required to segment complex craniospinal volumes by as much as 58% for an experienced clinician without loss of accuracy.
As with a preceding investigation involving the segmentation of mandible structures, the results reported here suggest that VR can serve as a viable and even preferable option for medical image segmentation and 3D anatomical modeling in support of pre-treatment planning or any other medical 3D visualization needs. Virtual reality can be used to create both 3D anatomical models and measurements with a high degree of accuracy and efficiency compared to those produced with conventional desktop platforms.
Availability of Data and Materials
All datasets used in this study, including de-identified medical images, are available from the corresponding author upon request.
References
Lenchik L, Heacock L, Weaver AA, Boutin RD, Cook TS, Itri J, et al. Automated Segmentation of Tissues Using CT and MRI: A Systematic Review. Acad Radiol 2019;26:1695–706. https://doi.org/10.1016/j.acra.2019.07.006.
Gharleghi R, Chen N, Sowmya A, Beier S. Towards automated coronary artery segmentation: A systematic review. Comput Methods Programs Biomed 2022;225:. https://doi.org/10.1016/j.cmpb.2022.107015.
Angulakshmi M, Lakshmi Priya GG. Automated brain tumour segmentation techniques— A review. Int J Imaging Syst Technol 2017;27:66–77. https://doi.org/10.1002/ima.22211.
Sutherland J, Belec J, Sheikh A, Chepelev L, Althobaity W, Chow BJW, et al. Applying Modern Virtual and Augmented Reality Technologies to Medical Images and Models. J Digit Imaging 2018;32:38–53. https://doi.org/10.1007/s10278-018-0122-7.
Williams CL, Kovtun KA. The Future of Virtual Reality in Radiation Oncology. Int J Radiat Oncol Biol Phys 2018;102:1162–4. https://doi.org/10.1016/j.ijrobp.2018.07.2011.
No Title. n.d. URL: https://www.realizemed.com/.
Chen C, Yarmand M, Singh V, Sherer M V., Murphy JD, Zhang Y, et al. VRContour: Bringing Contour Delineations of Medical Structures Into Virtual Reality. Proc - 2022 IEEE Int Symp Mix Augment Reality, ISMAR 2022 2022:64–73.https://doi.org/10.1109/ISMAR55827.2022.00020
Chen C, Yarmand M, Singh V, Sherer M V., Murphy JD, Zhang Y, et al. Exploring Needs and Design Opportunities for Virtual Reality-based Contour Delineations of Medical Structures. EICS 2022 - Companion 2022 ACM SIGCHI Symp Eng Interact Comput Syst 2022:19–25. https://doi.org/10.1145/3531706.3536456.
Chris Williams KK. www.dicomvr.com. 2023. URL: www.dicomvr.com.
Gruber LJ, Egger J, Bönsch A, Kraeima J, Ulbrich M, van den Bosch V, et al. Accuracy and Precision of Mandible Segmentation and Its Clinical Implications: Virtual Reality, Desktop Screen and Artificial Intelligence. Expert Syst Appl 2024;239:. https://doi.org/10.1016/j.eswa.2023.122275.
Anik AA, Xavier BA, Hansmann J, Ansong E, Chen J, Zhao L, et al. Accuracy and Reproducibility of Linear and Angular Measurements in Virtual Reality: a Validation Study. J Digit Imaging 2019;33:111–20. https://doi.org/10.1007/s10278-019-00259-3.
Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30:1323–41. https://doi.org/10.1016/j.mri.2012.05.001.
Dice LR. Measures of the Amount of Ecologic Association Between Species Author ( s ): Lee R . Dice Published by : Ecological Society of America Stable URL : http://www.jstor.org/stable/1932409. Ecology 1945;26:297–302.
Ajithkumar T, Horan G, Padovani L, Thorp N, Timmermann B, Alapetite C, et al. SIOPE – Brain tumor group consensus guideline on craniospinal target volume delineation for high-precision radiotherapy. Radiother Oncol 2018;128:192–7. https://doi.org/10.1016/j.radonc.2018.04.016.
Author information
Authors and Affiliations
Contributions
JB, image processing and modeling, image preparation, data analysis, and manuscript preparation and editing. JS and DL, image processing and modeling, image preparation, data analysis, and manuscript editing. MV, image processing and data collection analysis. KR, TF, EPDO, RG, MS, JW, MM, DG, and RA, study design and manuscript editing. VN, study design, data collection, analysis, and manuscript editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Research ethics board approval was obtained prior to the collection and analysis of clinical medical images in this investigation.
Consent for Publication
All authors have approved the manuscript for submission.
Competing Interests
JS and DL report a relationship with Realize Medical Inc. that includes board membership, employment, and equity ownership. They also have a patent pending as inventors on the underlying virtual reality technology used in this investigation. KR, JW, and RA are advisors for Realize Medical Inc. No other authors have conflicts of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Belec, J., Sutherland, J., Volpini, M. et al. A Pilot Clinical and Technical Validation of an Immersive Virtual Reality Platform for 3D Anatomical Modeling and Contouring in Support of Surgical and Radiation Oncology Treatment Planning. J Digit Imaging. Inform. med. (2024). https://doi.org/10.1007/s10278-024-01048-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10278-024-01048-3