Abstract
Deep learning (DL) has recently attracted attention for data processing in positron emission tomography (PET). Attenuation correction (AC) without computed tomography (CT) data is one of the interests. Here, we present, to our knowledge, the first attempt to generate an attenuation map of the human head via Sim2Real DL-based tissue composition estimation from model training using only the simulated PET dataset. The DL model accepts a two-dimensional non-attenuation-corrected PET image as input and outputs a four-channel tissue-composition map of soft tissue, bone, cavity, and background. Then, an attenuation map is generated by a linear combination of the tissue composition maps and, finally, used as input for scatter+random estimation and as an initial estimate for attenuation map reconstruction by the maximum likelihood attenuation correction factor (MLACF), i.e., the DL estimate is refined by the MLACF. Preliminary results using clinical brain PET data showed that the proposed DL model tended to estimate anatomical details inaccurately, especially in the neck-side slices. However, it succeeded in estimating overall anatomical structures, and the PET quantitative accuracy with DL-based AC was comparable to that with CT-based AC. Thus, the proposed DL-based approach combined with the MLACF is also a promising CT-less AC approach.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Positron emission tomography (PET) [1] is a powerful imaging modality used in clinical medicine and medical research. Other equally powerful clinical imaging modalities include computed tomography (CT) and magnetic resonance (MR) imaging. Attenuation correction (AC) [2] and scatter correction (SC) [3] are two essential components of PET data processing for quantitative PET imaging. The accuracy of the attenuation map directly affects the correction accuracy, and tremendous efforts have been made to improve the accuracy of PET data correction [4].
In combined PET-CT scanners, an acquired CT image is used to generate an attenuation map by applying simple piecewise linear scaling to CT values [5]. In combined PET-MR scanners, an attenuation map is usually generated by segmenting an acquired MR image into several regions and assigning known attenuation coefficients to the regions [6]. Before the advent of combined PET-CT, a transmission source, such as a 137-Cs point source and 68-Ge-68-Ga rod source, rotates around the subject, and an attenuation map is reconstructed from the acquired transmission data [7]. The breakthrough discovery in 2012 [8] that time-of-flight (TOF) PET emission data can determine attenuation factors except for a global scale factor triggered the development of joint reconstruction algorithm of activity and attenuation [9], i.e., combined with some scaling compensation method, attenuation map can be reconstructed from an acquired emission data without using any CT/MR data or transmission data. Bal et al. [10] developed a joint reconstruction algorithm combined with the activity domain scaling compensation method and showed using clinical brain PET data its good correlation with CT-based attenuation correction (CT-AC). In a recent dedicated head TOF-PET scanner [11], an attenuation map is reconstructed using a joint reconstruction algorithm combined with an attenuation domain scaling compensation method [12, 13].
Deep learning (DL) has recently attracted attention for data processing in PET [14,15,16], and attenuation correction (AC) without CT data is one of the interests. As listed in the recent review paper [17], there are four DL-based approaches to generate attenuation maps in terms of the input–output relation: (1) MR to pseudo-CT attenuation map, (2) non-attenuation-corrected (NAC) PET to pseudo-CT, (3) joint reconstruction output to CT attenuation map, and (4) NAC PET to corrected PET, i.e., (1) is the MR-based approach, and the others (2)–(4) are PET-based approaches. The pseudo-CT can be converted to an attenuation map by the scaling method mentioned above. The limitation of each approach is as follows [17]: the MR-based approach requires an MR image, and its performance depends on input MR image artifacts and PET-MR registration error. Although the PET-based approaches are free from such limitations due to their self-closed nature, common limitations of the PET-based approaches (2) and (4) is limited bone and air cavity delineation due to poor anatomical information on input NAC PET images. The PET-based approach (3) requires additional image reconstruction time, and its applicability is limited to the TOF-PET system.
In this paper, we present a new method employing tissue composition (TC) estimation to generate attenuation maps for the human head (Fig. 1), a DL-based approach that differs from the existing four approaches. Our research started from an idea that image soft (fuzzy) segmentation, which estimates the composition (mixture) ratio of multiple components in a single pixel, can handle the situation in PET imaging that multiple types of tissues (e.g., soft tissue, bone, and cavity) mixed within a single voxel of attenuation map. Conventional hard segmentation, in which a single label is assigned to each voxel, assumes that a single voxel consists of a single type of tissue; however, as voxel size gets larger, such assumption no longer holds, and soft segmentation is reasonable in such situation. Here, we state that our research motivation is not to overcome the limitations of the existing DL-based attenuation map generation approaches but to combine soft segmentation DL processing and joint image reconstruction. Then our approach shares the same limitations with the existing PET-based approaches.
Anatomical components of the human head can be roughly partitioned into the following three parts: soft tissue (including muscle, fat, and brain), bone, and cavity. In addition, the approximate attenuation coefficients of these three regions are known because segmentation-based MR-AC methods use these coefficients [18]. An attenuation map can be generated by a linear combination of the estimated TC map, as shown in Fig. 1. Although it is straightforward to use the DL estimate of the attenuation map directly for AC, we implement it as an initial attenuation map for reconstruction by the maximum likelihood attenuation correction factor (MLACF) [19], and thus, the DL estimate is refined by the MLACF. This indirect approach enables the integration of DL processing with existing data processing in our dedicated head TOF-PET scanner [13] released in 2021 in Japan.
The feasibility of our new DL-based AC approach is demonstrated using a limited number of clinical brain PET data, in which the accuracy of DL-based AC is compared with that of CT-AC. The results indicate that the proposed approach can be a promising fifth approach for DL-based PET AC.
Method
Proposed DL Model
Our proposed method is based on modeling an attenuation map of the human head as a linear combination of four TC maps of soft tissue, bone, cavity, and background (Fig. 1). The voxel value of the TC map represents the volumetric mixture ratio of a certain tissue in the voxel. Therefore, the sum of four TC values always equals one. We believe that no more fine partitioning is needed to achieve clinically acceptable quantitative accuracy inside a brain region.
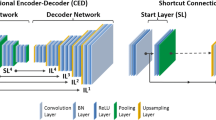
The proposed DL model accepts a two-dimensional intensity-normalized NAC PET image as input and outputs a four-channel TC map. This DL task can be seen as an instance of the semantic soft segmentation [20]. A modified U-net [21] was employed as the neural network structure (Fig. 2). Specifically, the input NAC image was encoded repeatedly to lower dimensional feature space via the blocks consisting of stride-1 convolution, batch normalization (BN), and leaky ReLU (LReLU) followed by downsampling by stride-2 convolution and then, to reach TC maps, decoded repeatedly to higher dimensional feature space via the blocks consisting of bilinear upsampling, stride-1 convolution, BN, and LReLU, with concatenating the intermediate feature maps in encoding stage to that of decoding stage. The soft-max activation in the output layer, which consists of similar building blocks used in the encoding and decoding stages, inherently guarantees the aforementioned sum-one constraint of the TC values.
Head position along the axial direction in the field of view may vary from patient to patient. If 3-D U-net was employed, such positional variations of the head must be considered in training dataset preparation unless some positional-invariance mechanism is introduced into the network architecture of the DL model. Then, employing 2-D U-net is a simple way to free from the consideration.
An attenuation map can be generated by a linear combination of the estimated four TC maps with known attenuation coefficients of soft tissue, bone, cavity, and background. The approximate attenuation coefficients for soft tissue, bone, and cavity are 0.096 mm−1, 0.0146 mm−1, and 0.0 mm−1, respectively [18].
In our approach, if needed, a final attenuation map can be tuned quantitatively by adjusting those attenuation coefficients or by post-processing the estimated TC maps (e.g., thresholding small TC values and smoothing the four TC maps in different manners). This kind of attenuation map tuning is impossible in existing DL-based approaches, which directly estimate the pseudo-CT values or attenuation coefficients. Therefore, this tunable feature distinguishes our indirect approach from existing direct approaches.
Reconstruction Flow
In our reconstruction flow (Fig. 3), the DL estimate of the attenuation map is used as input for scatter + random (S + R) estimation and as an initial estimate for attenuation map reconstruction by the MLACF, i.e., the DL estimate is refined by the MLACF. If the DL estimate is highly reliable, this refinement is not required. Processing with DL-based attenuation map generation was introduced into the existing reconstruction flow of our dedicated head TOF-PET scanner [13]. In the scanner, a uniform attenuation map without bone and cavity regions, derived from the NAC image is currently used as input for S + R estimation and as an initial estimate for attenuation map reconstruction. The use of a uniform attenuation map is not optimal but easy to implement and provides clinically acceptable results, as reported in a clinical study [22]. In short, we replaced the uniform attenuation map in the existing reconstruction flow with the DL estimate.
The accuracy of S + R estimation and attenuation map reconstruction can be improved by using the DL estimate, but this aspect was not investigated in detail in this study. In addition, alternating iteration of S + R estimation and attenuation map reconstruction, which is expected to provide better PET quantification, was not tested to simplify the reconstruction flow.
Dataset for Training
The DL model for TC estimation was trained using only the simulated dataset generated by an in-house analytical PET simulation tool. This Sim2Real approach of applying AI models trained only on simulated data to real-world data is uncommon in DL-based PET data processing and is completely distinct from previous DL-based AC methods using real-world (i.e., clinical) datasets for model training [23,24,25]. Although it is straightforward to train AI models using real-world data, it is beneficial to be able to build practical AI models using only simulated data, because it is not easy for companies to access large amounts of clinical data and that is a major barrier for companies to start developing AI for medical data. In addition, if realistic simulation data can be prepared, AI for new or rare cases (e.g., new radiotracers, rare diseases) can be developed without waiting for large amounts of real data to be accumulated.
Using the BrainWeb dataset [26], we generated a variety of activity and attenuation maps as input for PET simulation and the corresponding TC maps. The BrainWeb dataset includes a collection of high-resolution, three-dimensional (3-D) human head images of anatomical labels, which are derived from MR images of 20 adult subjects [26]. Seventeen subjects were used in the training step (12 for training and 5 for validation). We manually modified the anatomical labels. Specifically, labels of cavities corresponding to sinuses around the nose, cavities mimicking mastoid cells, and skin were added or corrected.
The TC maps were generated by conceptually binning the high-resolution labels of 0.5-mm isotropic voxels to the low-resolution labels of 4.5-mm isotropic voxels, i.e., the TC values in each low-resolution voxel are computed by binning 9 × 9 × 9 voxels. Here, 4.5 mm corresponds to the voxel size of the simulated NAC image.
Activity and attenuation maps were generated by assigning relative activity values and attenuation coefficients to each anatomical region. Here, to simulate 18F-fluorodeoxyglucose (FDG) and amyloid-imaging radiotracers (e.g., 18F-Flutemetamol, 18F-Florbetapir, and 18F-Florbetaben), we generated seven activity patterns with different gray matter (GM) to white matter (WM) concentration ratios in the range of 1:8 to 8:1 (Fig. 4).
The diameter and axial length of a simulated cylindrical non-TOF scanner were 300 mm and 180 mm, respectively, and the sinogram size was 128 [radial bins] × 64 [angular bins] × 452 [ring pairs]. The radial bin pitch was 2.0 mm, and the ring pitch was 4.0 mm. The scatter sinogram computed by the single scatter simulation [27] and the uniform random sinogram were added to the forward projection true sinogram after adjusting their number of counts. Scatter fractions were set to 30% and 40%, and the random fraction was set to 30%. The simulated numbers of prompt counts were 16 M, 32 M, and 64 M. To simulate the finite spatial resolution of the PET scanner, noise-free prompt sinograms were blurred by a Gaussian filter. Noisy prompt sinograms of all simulation settings of all subjects were reconstructed using 3-D ordered-subsets expectation–maximization (OS-EM) [28] without AC and SC at the 4.5-mm isotropic voxel with a matrix of 64 × 64 × 64.
To ensure the robustness of the DL model, in addition to general data augmentation by the affine transformations (flip, rotation, shift, and scaling), global intensity scaling and local intensity scaling for the background and skin regions were applied independently after intensity normalization. Finally, we prepared approximately 660,000 slices of NAC images for each GM-to-WM ratio for training. Examples of simulated NAC images of 16-M prompts are shown in Fig. 5.
Model Training
We trained two DL models: the GM model for GM-dominant radiotracers and the WM model for WM-dominant radiotracers. Specifically, the GM and WM models were trained using the simulated dataset of GM-to-WM ratios in the range of 8:1–1:1 and 1:1–1:8, respectively. Although it is possible to train a single general-purpose model with a mixed dataset of PET tracers [29], we trained multiple dedicated models to minimize the scope of influence when upgrading the models.
The cross-entropy loss was optimized using a gradient-based optimization algorithm (ADAM) [30] with a learning rate of 0.001, which was reduced by half if the validation loss did not decrease after ten training epochs. The mini-batch size was 1100, and the number of epochs was 400. The DL model was trained using the Keras package (ver.2.40, https://keras.io/) with the TensorFlow libraries (ver.2.30) [31] as computing backend. NVIDIA DGX Station (OS: Ubuntu 18.04.3 LTS, CPU: Intel Xeon E5-2698 v4 2.20 GHz, System memory: 256 GB, GPU: NVIDIA Tesla V100, GPU memory: 64 GB) was used for model training.
Dataset for Preliminary Evaluation
To demonstrate the feasibility of the proposed DL-based AC approach, three patients’ data were used: two patients of 18F-Flutemetamol (one is amyloid positive, and the other is amyloid negative) and one patient of 18F-FDG. These data were obtained in a past clinical study using our dedicated head TOF-PET scanner [22]. In the reconstruction flow, the NAC image was reconstructed at the 4.4-mm isotropic voxel with a matrix of 60 × 60 × 37 using 3-D OS-EM. After padding the in-plane matrix size to 64 × 64 and intensity normalization, each slice of the NAC image was inputted to the DL model. The GM model was applied to the 18F-FDG data, and the WM model was applied to the two 18F-Flutemetamol data.
PET Quantitative Evaluation
PET quantitative evaluation was performed for the attenuation and scatter-corrected images computed through the reconstruction flow mentioned above (Fig. 3). Final 3-D reconstruction was performed at the 2.2-mm isotropic voxel with a matrix of 120 × 120 × 74 using the list-mode dynamic row-action maximum-likelihood algorithm [32] with the post-median filter. As a comparison, reconstructions with CT-AC [5] were performed using CT images of the same subjects acquired during the same clinical study. Prior to the reconstructions, CT images were registered to the PET image coordinate system by using the 3-D rigid registration method based on the optimization of mutual information [33]. After applying anatomical standardization to the reconstructed images using SPM12 (https://www.fil.ion.ucl.ac.uk/spm/software/spm12/), the mean voxel values in ten anatomical regions (frontal lobe, temporal lobe, parietal lobe, occipital lobe, cerebellum, brain stem, striatum, thalamus, cingulate gyrus, and white matter) and the relative error between the proposed DL-based AC and CT-AC were computed using CT-AC as the baseline.
Results
The estimated TC and attenuation maps before and after MLACF refinement are shown with the corresponding input NAC images and CT-based attenuation maps in Figs. 6, 7, and 8. In the figures, DL and DL + MLACF mean before and after MLACF refinement, respectively. The CT-based attenuation maps show that anatomical structures in the neck-side slices are more complex than that in the parietal-side slices, and the proposed DL-based method tends to estimate anatomical details inaccurately, especially in the neck-side slices. In 18F-Flutemetamol cases (Figs. 7 and 8), the attenuation coefficient of 0.0160 mm−1 was used for bone in the linear combination process to compensate underestimation of the bone TC value. The reconstructed PET images with DL-based AC and CT-AC and their relative error maps are shown in Fig. 9, and the corresponding attenuation maps and their error (subtraction) maps are shown in Fig. 10. In Fig. 9, the relative activity errors in most brain regions are less than 5.0%; however, the other regions of bones and cavities have errors larger than 10%. As shown in Fig. 10, bones and cavities correspond to the regions having large attenuation errors. Specifically, attenuation coefficients of bones and cavities tend to be underestimated and overestimated, respectively. The joint histograms of the reconstructed activity values with the DL-based AC and CT-AC are shown in Fig. 11. The histogram bin width was unified in the two 18F-Flutemetamol cases but differs from that of the 18F-FDG case because their range of activity values is different. The joint histograms indicate that overall activity correlations between the DL-based AC and CT-AC are high; however, the reconstructed activity values with the DL-based AC tend to be underestimated at high activity voxels, and the relative errors are larger for low activity voxels than for high activity voxels. These trends are shared by both the radiotracers.
Representative slices of the estimated TC and attenuation maps before and after MLACF refinement with corresponding input NAC images and CT-based attenuation maps for the 18F-Flutemetamol case (amyloid negative). The arrows indicate that overestimation of the attenuation coefficients in the nasal cavities and mastoid cells and underestimation of the skull thickness in DL can be improved through the MLACF in DL + MLACF
Joint histograms of reconstructed activity values with DL-based AC (DL + MLACF) and CT-AC of three subjects. The dot lines indicate the relative error range of 5% and 10%. The histogram bin width was unified in the two 18F-Flutemetamol cases (b–c) but differs from that of the 18F-FDG case (a) because their range of activity values is different
Axial slices including nasal cavities are separately shown in Fig. 12. Attenuation error maps in Fig. 12 also show that low attenuation regions such as nasal cavities and mastoid cells have large positive errors and, by comparing them with the activity error maps in Fig. 9, it is apparent that the large attenuation errors of such regions were expansively propagated to the PET images.
The relative errors of mean voxel values at each anatomical region between the proposed DL-based AC (DL + MLACF) and CT-AC are summarized in Table 1. In this table, error values without MLACF refinement (i.e., DL-estimated attenuation map was directly used in the list-mode reconstruction) were also included. The comparison between the DL + MLACF and the DL shows that the MLACF refinement resulted in a decrease in reconstructed activity values. The bottom row of the table shows the counts of regions having smaller absolute errors. There are 30 regions in total (ten regions per patient by three patients) without distinguishing between patients, and the DL + MLACF has smaller errors in eight regions, and the DL has smaller errors in the other 22 regions. This means that, in our study using the three patients’ data, we could not demonstrate the positive effects of post-processing the DL-estimated attenuation map by the MLACF on PET quantification.
The computation time of our DL inference was less than 1.0 s per patient (i.e., not per slice) in our specific computation environment. The computational complexity of our DL inference would be comparable or lower to that of the direct generation methods because we employed ordinary simple 2-D U-net architecture; however, our reconstruction flow (Fig. 3) contains attenuation map reconstruction step by MLACF then its computation time increases overall computation time. This is the same as the existing joint reconstruction-based DL approach mentioned in the introduction. In principle, the computation time of list-mode reconstruction is proportional to the number of measured coincidence events, and the overall processing time using GPU (NVIDIA Quadro RTX 5000) was approximately 10 min per patient on average.
Discussion
In this study, we developed a new method based on TC estimation to generate Sim2Real DL-based attenuation maps for the human head. TC estimation is an instance of semantic soft segmentation. The attenuation map was reconstructed by the MLACF using the DL estimate of the attenuation map (i.e., a linear combination of the TC maps) as the initial estimate. Here, the DL models converting the NAC PET image to the four-channel TC map were trained with only the simulated dataset computed by using the in-house analytical PET simulation tool and the open database of high-resolution human head images of anatomical labels. Our Sim2Real approach is distinct from previous DL-based AC methods using clinical datasets for model training.
According to the attenuation maps shown in Figs. 6, 7, 8, 10, and 12, the proposed DL-based method tends to estimate anatomical details inaccurately, especially in the neck-side slices. However, a visual inspection indicated that the head shape and overall anatomical structure of soft tissues and bone were estimated well. There are two reasons for the inaccuracy in the neck-side slices. First, the anatomical structures of the cavity and bone in the neck-side slices vary widely from individual to individual, and our training dataset derived from 12 subjects would not capture the variety of structures. Second, the axial coverage of the BrainWeb dataset is limited to 181 mm, which covers the whole brain but not the whole head. Therefore, the neck-side slices including oral cavities are completely missed and the neck-side slices including nasal cavities are partially missed in the training dataset. To remedy those problems, a larger number of whole-head MR images should be collected and segmented, although this task is not easy in terms of human, time, and financial resources. In any case, the results showed the practicality of the proposed method in the brain region, as we expected.
According to the TC maps shown in Figs. 6, 7, and 8, the bone TC value of the 18F-Flutemetamol cases tends to be smaller than that of the 18F-FDG case. Therefore, the bone region is obscure on the attenuation map, which negatively affects PET quantification (omitted in this paper). The larger noise in the input NAC images of 18F-Flutemetamol cases may cause the degradation.
To compensate for the degradation in the 18F-Flutemetamol cases, we adjusted the attenuation coefficient of bone used in the linear combination process. Specifically, to compensate for the underestimation of the bone TC value, the excessive attenuation coefficient of 0.0160 mm−1 was used for bone instead of 0.0146 mm−1, which was used in the 18F-FDG case. Although this kind of manual parameter adjustment is not preferred, our attenuation map generation approach based on TC estimation allows this tuning to obtain the preferred output.
The joint histograms of the PET images shown in Fig. 11 indicate that overall activity correlations between the DL-based AC and CT-AC are high and, as shown in Table 1, the relative quantification errors of all the anatomical regions are less than 5.0% in both the DL + MLACF and the DL. According to the relative error maps shown in Fig. 9, although some regions have relatively large errors, the relative errors in most brain regions are less than 5.0%. This quantification error is worse than that of the previous DL-based head AC method [20], in which the average errors are less than 1% in most brain regions. According to Fig. 9, the relative errors outside of the brain region are locally large, especially inside the low attenuation regions and surrounding areas. These large errors likely originate in the overestimation of the attenuation coefficients of nasal and oral cavities in the attenuation maps (see Figs. 10 and 12). In addition to cavity regions, attenuation coefficients in bone regions were underestimated. Attenuation errors in bone regions generally affect PET quantification of not only the corresponding bone regions but also its neighborhood regions such as cerebral cortex regions. As shown in Table 1, the reconstructed activity values of the cerebral cortex regions (frontal lobe, temporal lobe, parietal lobe, and occipital lobe) were underestimated in both the DL + MLACF and the DL in comparison to the CT-AC. The main reason for this underestimation may be due to the deviation of the attenuation coefficients in the CT-AC and the template attenuation coefficients we assigned in this study. As discussed afterwards, although PET quantification performance was slightly degraded with the addition of the MLACF in comparison between the DL + MLACF and the DL, and there would be room for improving the DL model, we believe that reasonable quantitative performance was obtained with the proposed different types of the approach of combining DL processing and joint image reconstruction.
According to Fig. 8, the MLACF can compensate visually for the inaccuracy of the DL estimate of attenuation maps. Specifically, as indicated by the arrows in Fig. 8, the overestimation of the attenuation coefficients in the nasal cavities and mastoid cells and the underestimation of the skull thickness were improved through the MLACF. On the other hand, as shown in Figs. 6, 7, and 8, the attenuation coefficients in the bone regions tend to decrease through the MLACF, and the bone regions tend to be visually obscured. As mentioned above, the trained DL model estimates anatomical details inaccurately, and then we believe that the MLACF using the DL estimate as its initial estimate would be inherently required. However, in this study, we could not demonstrate quantitatively that the DL + MLACF outperforms the DL without MLACF refinement. According to Table 1, the addition of the MLACF resulted in a decrease in reconstructed activity values. At least, the decrease of the attenuation coefficients in the skull through the MLACF directly resulted in the decrease of reconstructed activity values of cerebral cortex regions because the regions are adjacent to the skull. In future studies, we should clarify the statistically quantitative performance of the proposed method and try to adjust processing parameters including the template attenuation coefficients if needed.
This study has several limitations. First, the number of subjects in the evaluation is minimal. Therefore, further investigations should be conducted with a larger number of subjects in the 18F-FDG and 18F-Flutemetamol studies, with an emphasis on the versatility of the trained DL models and statistical difference between the proposed DL-based AC and CT-AC. In such investigations, the robustness of the trained DL model against variation of input NAC image in terms of anatomical and functional spatial patterns and noise level would be of major interest. The noise level could fluctuate from patient to patient depending on administration dose, uptake time, data acquisition time, etc. Second, the versatility of the excessive attenuation coefficient of 0.0160 mm−1 used for bone in the 18F-Flutemetamol cases is unknown and should be verified and/or optimized by using a dataset comprising a large number of subjects. Third, the applicability of the trained GM and WM models to other radiotracers is unknown. Although we expect that the trained DL models work to some extent with other radiotracers exhibiting an activity distribution close to that of 18F-FDG and 18F-Flutemetamol, fine-tuning of the trained DL models using the simulated dataset of the target radiotracers would be required.
Conclusion
Here, we present, to our knowledge, the first attempt to generate attenuation maps of the human head via Sim2Real DL-based tissue composition estimation from model training using only the simulated PET dataset. The proposed DL model trained using only the simulated dataset tended to estimate anatomical details inaccurately, especially in the neck-side slices. However, it succeeded in estimating overall anatomical structures, and the accuracy of DL-based AC was comparable to that of CT-AC. Although the number of clinical PET data and kind of radiotracers were limited in the evaluation, we believe that the proposed DL-based attenuation map generation approach combined with MLACF is also a promising CT-less AC approach.
References
Phelps ME. PET: Molecular imaging and its biological applications. New York: Springer; 2012.
Habib Z, Montandon M, Alavi A. Advances in attenuation correction techniques in PET. PET Clin. 2007;2:191–217. https://doi.org/10.1016/j.cpet.2007.12.002.
Habib Z, Montandon M. Scatter compensation techniques in PET. PET Clin. 2007;2:219–234. https://doi.org/10.1016/j.cpet.2007.10.003.
Mohammad D, Jiang X, Schäfers K. Correction techniques in emission tomography. Boca Raton: CRC Press; 2012.
Burger C, Goerres G, Schoenes S, Buck A, Lonn AHR, Von Schulthess GK. PET attenuation coefficients from CT images: experimental evaluation of the transformation of CT into PET 511-keV attenuation coefficients. Eur J Nucl Med Mol Imaging. 2002;29:922–927. https://doi.org/10.1007/s00259-002-0796-3.
Ladefoged CN, Law I, Anazoda U, Lawrence KS, Izquierdo-Garcia D, Catana C, et al. A multi-centre evaluation of eleven clinically feasible brain PET/MRI attenuation correction techniques using a large cohort of patients. Neuroimage. 2017;147:346–359. https://doi.org/10.1016/j.neuroimage.2016.12.010.
Bailey DL. Transmission scanning in emission tomography. Eur J Nucl Med. 1998;25:774–787. https://doi.org/10.1007/s002590050282.
Defrise M, Ahmadreza R, Nuyts J. Time-of-flight PET data determine the attenuation sinogram up to a constant. Phys Med Biol. 2012;57.4:885. https://doi.org/10.1088/0031-9155/57/4/885
Berker Y, Li Y. Attenuation correction in emission tomography using the emission data—a review. Med Phys. 2016;43.2:807-832. https://doi.org/10.1118/1.4938264
Bal H, Panin VY, Platsch G, Defrise M, Hayden G, Hutton C, Serrano B, Paulmier B, Casey ME. Evaluation of MLACF based calculated attenuation brain PET imaging for FDG patient studies. Phys Med Biol. 2017;62.7:2542. https://doi.org/10.1088/1361-6560/aa5e99
Morimoto-Ishikawa D, Hanaoka K, Watanabe S, Yamada T, Yamakawa Y, Minagawa S, Tekenouchi S, Ohtani A, Mizuta T, Kaida H, Ishii K. Evaluation of the performance of a high-resolution time-of-flight PET system dedicated to the head and breast according to NEMA NU 2-2012 standard. EJNMMI Phys. 2022;9:88. https://doi.org/10.1186/s40658-022-00518-3.
Kobayashi T, Kitamura K. A solution for scaling problem in joint estimation of activity and attenuation. 2017 IEEE NSS/MIC, Atlanta, GA, USA. 2017:1–5. https://doi.org/10.1109/NSSMIC.2017.8532856.
Mizuta T, Kobayashi T, Yamakawa Y, Hanaoka K, Watanabe S, Morimoto-Ishikawa D, Yamada T, Kaida H, Ishii K. Initial evaluation of a new maximum-likelihood attenuation correction factor-based attenuation correction for time-of-flight brain PET. Ann Nucl Med. 2022;36:420–426. https://doi.org/10.1007/s12149-022-01721-z.
Gong K, Berg E, Cherry S, Qi J. Machine learning in PET: from photon detection to quantitative image reconstruction. Proc IEEE. 2019;108:51–68. https://doi.org/10.1109/JPROC.2019.2936809.
Wang T, Lei Y, Curran WJ, Liu T, Nye JA, Yang X. Machine learning in quantitative PET: A review of attenuation correction and low-count image reconstruction methods. Phys Med. 2020;76:294–306. https://doi.org/10.1016/j.ejmp.2020.07.028.
Arabi H, Allaf AA, Sanaat A, Shiri I, Zaidi H. The promise of artificial intelligence and deep learning in PET and SPECT imaging. Phys Med. 2021;83:122–137. https://doi.org/10.1016/j.ejmp.2021.03.008.
Lee JS. A review of deep-learning-based approaches for attenuation correction in positron emission tomography. IEEE Trans Radiat Plasma Med Sci. 2020;5:160–184. https://doi.org/10.1109/TRPMS.2020.3009269.
Wagenknecht G, Kaiser HJ, Mottaghy MF, Herzog H. MRI for attenuation correction in PET: methods and challenges. MAGMA. 2013;26:99–113. https://doi.org/10.1007/s10334-012-0353-4.
Ahmadreza R, Defrise M, Nuyts J. ML-reconstruction for TOF-PET with simultaneous estimation of the attenuation factors. IEEE Trans Med Imaging. 2014;33:1563–1572. https://doi.org/10.1109/TMI.2014.2318175.
Aksoy Y, Oh T, Paris S, Pollefeys M, Matusik W. Semantic soft segmentation. ACM Trans Graph. 2018;37:1-13. https://doi.org/10.1145/3197517.3201275.
Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical Image Computing and Computer-Assisted Intervention. MICCAI 2015. Lecture Notes in Computer Science, vol. 9351. Springer; 2015. https://doi.org/10.1007/978-3-319-24574-4_28
Ishii K, Hanaoka K, Watanabe S, Morimoto-Ishikawa D, Yamada T, Kaida H, Yamakawa Y, Minagawa S, Takenouchi S, Ohtani A, Mizuta T. High-resolution silicon photomultiplier time-of-flight dedicated head PET system for clinical brain studies. J Nucl Med. 2023;64:153–158. https://doi.org/10.2967/jnumed.122.264080.
Liu F, Jang H, Kijowski R, Zhao G, Bradshaw T, McMillan AB. A deep learning approach for 18F-FDG PET attenuation correction. EJNMMI Phys. 2018;5:1–15. https://doi.org/10.1186/s40658-018-0225-8.
Dong X, Wang T, Lei Y, Higgins K, Liu T, Curran WJ, Mai H, Nye JA, Yang X. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging. Phys Med Biol. 2019;64:215016. https://doi.org/10.1088/1361-6560/ab4eb7.
Armanious K, Hepp T, Küstner T, Dittman H, Nikolaou K, La Fougere C, Yang B, Gatidis S. Independent attenuation correction of whole body [18F] FDG-PET using a deep learning approach with Generative Adversarial Networks. EJNMMI Res. 2020;10:1–9. https://doi.org/10.1186/s13550-020-00644-y.
Aubert-Broche B, Griffin M, Pike GB, Evans AC, Collins DL. Twenty new digital brain phantoms for creation of validation image data bases. IEEE Trans Med Imaging. 2006;25:1410–1416. https://doi.org/10.1109/TMI.2006.883453.
Watson CC, Newport DF, Casey ME. A single scatter simulation technique for scatter correction in 3D PET. In: Grangeat P, Amans JL, editors. Three-dimensional image reconstruction in radiology and nuclear medicine. Computation imaging and vision, vol. 4. Dordrecht: Springer; 1996. pp. 255–268. https://doi.org/10.1007/978-94-015-8749-5_18.
Hudson HM, Larkin RS. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans Med Imaging. 1994;13:601–609. https://doi.org/10.1109/42.363108.
Hashimoto F, Ito M, Ote K, Isobe T, Okada H, Ouchi Y. Deep learning-based attenuation correction for brain PET with various radiotracers. Ann Nucl Med. 2021;35:691–701. https://doi.org/10.1007/s12149-021-01611-w.
Kingma DP, Ba AJ. A method for stochastic optimization. ArXiv e-prints [Internet]. 2014. Available from: http://arxiv.org/abs/1412.6980. Accessed 13 Nov 2023.
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. ArXiv e-prints [Internet]. 2016. Available from: http://arxiv.org/abs/1603.04467. Accessed 13 Nov 2023.
Nakayama T, Kudo H. Derivation and implementation of ordered-subsets algorithms for list-mode PET data. IEEE Nucl Sci Symp Conf Rec, Fajardo, PR, USA. 2005;4:1954. https://doi.org/10.1109/NSSMIC.2005.1596714
Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16:187–198. https://doi.org/10.1109/42.563664
Acknowledgements
This study continues from a previous research project (jRCTs052200055) funded by Shimadzu Corporation. The NAC images in Figs. 6, 7, and 8 are part of previous research data. We thank Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
TK and YS conceived the idea of the study. TK, YS, YY, and YT developed and implemented the computational algorithm. TK and YS designed and conducted the experiments to evaluate the performance of the algorithm. TK, YS, KH, SW, DMI, and TY contributed to the interpretation of the results. TK, YS, and TM drafted the original manuscript. TM, HK, and KI supervised the conduct of this study. All authors reviewed the manuscript draft and revised it.
Corresponding author
Ethics declarations
Ethics Approval
There are no procedures requiring ethics approval relevant to this article.
Consent to Participate
There are no procedures requiring informed consent relevant to this article.
Consent for Publication
All authors have approved the manuscript and agree with the submission.
Competing Interests
Tetsuya Kobayashi, Yui Shigeki, Yoshiyuki Yamakawa, Yumi Tsutsumida, and Tetsuro Mizuta are employees of Shimadzu Corp. There are no other potential conflicts of interest relevant to this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kobayashi, T., Shigeki, Y., Yamakawa, Y. et al. Generating PET Attenuation Maps via Sim2Real Deep Learning–Based Tissue Composition Estimation Combined with MLACF. J Digit Imaging. Inform. med. 37, 167–179 (2024). https://doi.org/10.1007/s10278-023-00902-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-023-00902-0