Abstract
Histone modification is an important epigenetic mechanism in eukaryotes. Histone acetyltransferase and deacetylase regulate histone acetylation levels antagonistically, leading to dynamic control of chromatin structure. One of the histone deacetylases, HDA6, is involved in gene silencing in the heterochromatin regions, chromocenter formation, and metabolic adaptation under drought stress. Although HDA6 plays an important role in chromatin control and response to drought stress, its intracellular localization has not been observed in detail. In this paper, we generated transformants expressing HDA6-GFP in the model plant, Arabidopsis thaliana, and the crops, rice, and cassava. We observed the localization of the fusion protein and showed that HDA6-GFP was expressed in the whole root and localized at the nucleus in Arabidopsis, rice, and cassava. Remarkably, HDA6-GFP clearly formed speckles that were actively colocalized with chromocenters in Arabidopsis root meristem. In contrast, such speckles were unlikely to be formed in rice or cassava. Because AtHDA6 directly binds to the acetate synthesis genes, which function in drought tolerance, we performed live imaging analyses to examine the cellular dynamics of pH in roots and the subnuclear dynamics of AtHDA6 responding to acetic acid treatment. The number of HDA6 speckles increased during drought stress, suggesting a role in contributing to drought stress tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In eukaryotes, DNA is organized in a higher order structure that achieves the mutually opposing requirements of compaction to package DNA in the nucleus and relaxation to allow replication and transcription to progress. The least compaction is achieved by forming a nucleosome, which is 147 bp of DNA wrapped around a histone octamer composed of histones H2A, H2B, H3, and H4. These histone proteins carry post-transcriptional modifications in their N-terminal tails. The modifications, including acetylation, methylation, phosphorylation, ubiquitylation, and sumoylation, regulate nucleosome concentration, leading to optimal chromatin structure such as euchromatin and heterochromatin.
The acetylation neutralizes the positive charge of lysine residues in the histone tail, which disrupts electrical interaction between DNA and histones and presumably loosens the compacted chromatin structure. The relaxation of chromatin structure enables accessibility to transcription factors and RNA polymerase, increasing the transcriptional activity (Bannister and Kouzarides 2011). Acetylation is dynamically regulated by histone acetyltransferase (HAT) and histone deacetylase (HDAC) (Allis and Jenuwein 2016; Zentner and Henikoff 2013). HAT catalyzes the transfer of an acetyl group from acetyl CoA to lysine residues, while HDAC removes acetylation from lysine residues and recovers the positive charge that leads to condensed chromatin structure.
Eighteen HDAC genes are known and are classified into three families in Arabidopsis thaliana: the RPD3/HDA1 superfamily, the Sirtuin family, and the HD2 family (Hollender and Liu 2008; Pandey et al. 2002). Twelve genes belong to the RPD3/HDA1 superfamily, which is subdivided into Classes I, II, and III. HDA6, 7, 9, and 19 are identified as Class I, AtHDA5, 15, and 18 are Class II, and HDA2 is Class III, based on sequence similarity (Pandey et al. 2002). AtHDA8, 10, 14, and 17 are members of the RPD3/HDA1 superfamily but remain unclassified (Pandey et al. 2002). It is reported that Class I HDAs play critical roles in many developmental processes and environmental stress responses. In particular, AtHDA6 is well studied as a global repressor involved in flowering, freezing tolerance, drought stress response, repression of embryonic properties, the jasmonate signaling pathway, and establishment of nucleolar dominance (Kim et al. 2012).
Most recently, Kim and colleagues clarified the crucial roles of AtHDA6 in drought tolerance. When plants are exposed to drought stress, acetate synthesis genes, including pyruvate decarboxylase PDC1 and acetaldehyde dehydrogenase ALDH2B7, are activated and acetate content is increased (Kim et al. 2017). The genes that function to acquire drought tolerance are acetylated using acetyl groups provided by the synthesized acetate, which enhances drought tolerance. AtHDA6 directly binds to the acetate synthesis genes and inhibits activation under normal conditions, while under drought stress AtHDA6 becomes disassociated from the acetate synthesis genes, leading to their activation (Kim et al. 2017).
AtHDA6 functions to organize heterochromatin, including transposable elements, centromere repeats, and ribosomal DNA repeats. Drastic reduction in DNA methylation in transposable elements, 45S rDNA, and 5S rDNA was detected in athda6 mutants (Earley et al. 2010; To et al. 2011; Vaillant et al. 2007). In the case of 5S rDNA, CNG methylation was reduced in the athda6 mutant, although the total amount of 5S rRNA was not affected (Vaillant et al. 2007). HDA6 is also involved in the organization of chromocenters, which are mainly composed of heterochromatin and can be visualized as DAPI-dense spots, because the number of chromocenters is smaller in the athda6 mutant than the wild-type (Vaillant et al. 2007).
Although HDA6 plays key roles in stress responses and chromatin organization, its subcellular localization patterns have not been observed in detail. In this study, we analyzed the localization of HDA6-GFP in Arabidopsis and the crops, rice, and cassava for live imaging.
Materials and methods
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana (L.) Heynh. ecotype Col-0), rice (Oryza sativa L. cv. Nipponbare), and cassava (Manihot esculenta Crantz cv. 60444) were used. The seeds of Arabidopsis and rice were sown on half-strength Murashige and Skoog (MS) medium with 1% sucrose and 1% gellan gum (pH 5.8 with KOH). After incubation at 4 °C for 24 h, the plants were grown at 22 °C for Arabidopsis and at 30 °C for rice, 16 h light/8 h dark for 5–8 days. Plantlets of the cassava cultivar were obtained from the in vitro germplasm collection of the International Center for Tropical Agriculture (CIAT), Cali, Colombia. The in vitro plantlets were maintained on MS medium with 20 g L−1 sucrose and 3 g L−1 Gelrite (pH 5.8 using KOH). The plantlets for experiments were grown at 28 °C, 16 h light/8 h dark until rooting.
Phylogenetic analysis
The amino acid sequences of HDACs from Arabidopsis, rice and cassava were obtained from TAIR10 (https://www.arabidopsis.org/), Oryza sativa v7.0 and Manihot esculenta v6.1 from the Joint Genome Institute (https://phytozome.jgi.doe.gov/pz/portal.html), respectively. A phylogenetic tree was constructed from full-length amino acid sequences based on the neighbor-joining method using CLC Genomics Workbench 11.0.1 software (https://www.qiagenbioinformatics.com/). Bootstrap probabilities for each node were calculated with 1000 replicates.
Generation of transformants expressing HDA6-GFP
HDA6 genomic sequences up to the end of the coding region were amplified by PCR using primers listed in Table S1. HDA6 genomic sequences of Arabidopsis (At5g63110), rice (Os08g25570) and cassava (Me14g061800) contained 524 bases, 2 kb, and 2 kb upstream of the start codon, respectively. The amplified fragments from Arabidopsis and rice were cloned into the pENTR/D-TOPO® plasmid using the pENTR/D-TOPO® Cloning Kit (Thermo Fisher Scientific, Waltham, MA, USA). The amplified fragment from cassava was cloned into the pDONR/Zeo plasmid using the BP clonase Kit (Thermo Fisher Scientific). The cloned fragments were transferred to the pGWB540 plasmid with Gateway® LR Clonase® II Enzyme Mix (Thermo Fisher Scientific) (Nakagawa et al. 2007). Using the constructs, Agrobacterium tumefaciens-mediated transformation of Arabidopsis, rice and cassava was performed as described previously (Sakamoto et al. 2018; Toki et al. 2006; Utsumi et al. 2017).
DAPI staining
For 4′,6-diamidino-2-phenylindole (DAPI) staining, roots of Arabidopsis, rice, and cassava were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 1 h at 25 °C. After washing with PBS, roots were stained with threefold diluted CyStain UV Precise P (Sysmex, Hyogo, Japan) containing DAPI for 15 min at 25 °C. Stained roots were mounted with 20%, 50%, and 100% TOMEI solution (TCI, Tokyo, Japan) (Hasegawa et al. 2016). Confocal images were obtained using a confocal laser scanning microscope (FV1200; Olympus, Tokyo, Japan) equipped with 405 nm, 473 nm, and 559 nm LD laser lines and a 100 × 1.40 numerical aperture (NA) oil immersion objective (UPlanSApo; Olympus), 60 × 1.40 NA oil immersion objective (PlanApo; Olympus), 40 × 1.30 NA oil immersion objective (UPlanFL; Olympus), and 20 × 0.75 dry objective (UPlanSApo; Olympus). The confocal images were analyzed using ImageJ ver.1.51 (http://rsb.info.nih.gov/ij/).
Fluorescence in situ hybridization (FISH) analysis
Arabidopsis roots expressing HDA6-GFP were fixed with 4% formaldehyde in PBS for 20 min at 25 °C. After washing in PBS, roots were treated with enzyme solution [1% Driselase (Sigma-Aldrich, St. Louis, Missouri, USA), 0.5% Cellulase R-10 (Yakult Pharmaceutical, Tokyo, Japan), 0.025% Pectolyase (Seishin Pharmaceutical, Saga, Japan) in PBS] for 45 s at 37 °C. Three to five root tips were arranged on a glass slide and squashed using a cover slip. HDA6-GFP signals were observed by confocal microscopy. After observation, slides were frozen with liquid N2 and the cover slip was removed. FISH analysis was performed as described previously (Hasegawa et al. 2018). In brief, slides were treated with RNase A, dehydrated with 70% and 100% ethanol then treated with 70% formamide for 2 min at 76 °C and immediately placed in 70% ethanol at − 20 °C for 5 min. Slides were treated with FISH probes synthesized by nick translation using a DIG nick translation mix (Roche, Basel, Switzerland) for 16 h at 37 °C. Primers for the probe synthesis were listed in Table S1. The probes were visualized using anti-digoxigenin-rhodamine Fab fragments (Roche). FISH signals were observed using confocal microscopy.
Microchannel device development
The polydimethylsiloxane (PDMS) microchannel device was constructed using a MEMS (microelectromechanical systems) soft lithography process as described elsewhere (Hida et al. 2015; Park et al. 2014). A negative master for the molding of PDMS was fabricated on a single crystal silicon substrate with an ultrathick photoresist (SU-8 3025, MicroChem, Newton, MA, USA) using a photolithographic process. A precursor of PDMS (Sylgard 184, Dow Corning, Midland, MI, USA) was applied to the microfabricated mold and then degassed in a vacuum chamber for at least 20 min. After degassing, the PDMS precursor was kept at 90 °C for 1 h then the coagulated PDMS structure was peeled off the mold. Finally, a through-hole for the outlet was formed with a punch, and the PDMS device was attached onto a glass substrate to form a microchannel.
pH monitoring
Arabidopsis thaliana accession Colombia (Col-0) was used for this study. A transgenic line of Apo-pHusion has been described previously (Gjetting et al. 2012). For live imaging of Apo-pHusion and pHusion in roots of Arabidopsis, we used an inverted microscope IX-83 (Olympus) equipped with a confocal scanning system CSU-X1 (Yokogawa Electric, Tokyo, Japan), 488 nm and 561 nm LD lasers (Melles Griot, Rochester, NY, USA), and sCMOS camera (Neo 5.5 sCMOS; Andor, Belfast, Abingdon, UK). Ratio images were prepared from acquired fluorescent images of the pH-sensitive enhanced green fluorescent protein (EGFP) and the pH-tolerant monomeric red fluorescent protein 1 (mRFP1) using MetaMorph (Molecular Devices, San Jose, California, USA). For 12 h time-lapse imaging, 7-day-old seedlings were transferred to the microchannel device and perfused with one-half MS medium with 1% sucrose (pH5.8 with KOH). For perfusion imaging, 7-day-old seedlings were transferred to the microchannel device and perfused with pH buffer (5 mM MES, 5 mM MOPS adjusted to pH 8 with 1 M Tris–HCl). The perfusion solution was changed under the microscope every 3 min to different buffers at pH 8–5 with the flow speed at 2 ml min−1.
Acetic acid, mannitol, and TSA treatment
For all treatments, 5- to 8-day-old seedlings were transferred to the microchannel device and were treated with half-strength MS medium containing 1% sucrose (pH 5.8 with KOH). After taking microscopic pictures at 0 h treatment, the solution was exchanged by perfusion with soil extract (pH 6.2) or acetic acid soil extract (pH 6.5) (made by soaking soil for 1 week in water or 10 mM acetic acid, respectively), acetic acid buffer (one-half MS medium with 1% sucrose, 5 mM MES, 5 mM MOPS and 5 mM acetic acid adjusted to pH 7.0 with 1 M Tris–HCl), half-strength MS medium containing 1% sucrose or 200 mM D-mannitol (Wako, Osaka, Japan) (pH 5.8 with KOH), and half-strength MS medium containing 1% sucrose or 1 µM TSA (Wako, Osaka, Japan) (pH 5.8 with KOH).
Immunostaining analysis
Arabidopsis roots expressing HDA6-GFP were fixed with 4% formaldehyde in PBS for 40 min at 25 °C. After washing in PBS, roots were treated with the enzyme solution (1% Driselase (Sigma-Aldrich, St. Louis, Missouri, USA), 0.5% Cellulase R-10 (Yakult Pharmaceutical, Tokyo, Japan), 0.025% Pectolyase (Seishin Pharmaceutical, Saga, Japan) in PBS) for 45 s at 37 °C. Three to five root tips were arranged on a glass slide and squashed using a cover slip. The slides were frozen with liquid N2 and the cover slip was removed. Immunostaining analysis was performed as described previously (Hirakawa et al. 2017). After the slides were dried, they were immersed in 0.5% (v/v) Triton-X 100/PBS for 15 min and then washed three times in PBS for 5 min each at 25 °C. Four per cent (w/v) bovine serum albumin (BSA)/PBS was dropped on the sample area. After 30 min, the 4% BSA/PBS solution was removed from the slide and then a mixture of primary antibody and 1% (w/v) BSA/PBS (1:100) was added. Anti‐H3K9ac (ab4441; abcam, Cambridge, UK) were used as the primary antibodies. The slides were incubated in a humid box at 4 °C overnight and then washed in PBS for 5 min. Next, a mixture of secondary antibody and 1% (w/v) BSA/PBS (1:500) was added. Goat Anti-Rabbit Alexa Fluor® 546 (Thermo Fisher Scientific) were used as the secondary antibodies. The slides were incubated at 37 °C for 60 min in the dark and then washed twice in PBS for 5 min. Finally, slides were immersed in 0.1 μg ml−1 DAPI/PBS for 5 min and then mounted under a cover glass with VECTASHIELD antifade mounting medium (Vector Laboratories, Burlingame, California, USA). Specimens were observed under a FV1200 confocal laser microscope equipped with a GaAsP detector.
Results
AtHDA6 in Arabidopsis root meristem cells has a unique expression pattern in nuclei
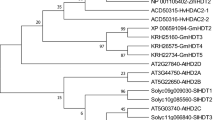
The Arabidopsis genome is known to possess 18 HDACs, comprising 12 RPD3/HDA1 genes, 4 HD2 genes, and 2 SIR2 genes (Hollender and Liu 2008). In contrast, our investigation of HDAC candidates having putative histone deacetylase domains found 18 and 15 HDACs in the rice and cassava genomes, respectively. The analysis revealed that both rice and cassava genomes contain all classes of RPD3/HDA1, HD2, and SIR2 homologues (Fig. 1a). HDA6 was conserved in both rice and cassava genomes: in both species the protein-coding sequences contained a histone deacetylase domain that showed high homology with AtHDA6 (77.3% and 82.3% in rice and cassava, respectively) (Fig. 1b).
Phylogenetic analysis of HDACs from Arabidopsis, rice, and cassava. a Neighbor-joining tree constructed from full-length amino acid sequences of HDAC homologues. Bootstrap probabilities from 1000 replications are represented at each node. Scale bar represents the number of substitutions per amino acid site. The enlarged window shows the cluster of HDAC proteins in Class I of RPD3/HDA1 family. b Full-length amino acid sequences of HDA6 in Arabidopsis, rice, and cassava. Black outline: histone deacetylase domain
It has been reported that the HDA6 speckle formed in the nucleus of Arabidopsis for immunostaining (Pontes et al. 2007). We analyzed the localization of HDA6-GFP in Arabidopsis, rice, and cassava for live imaging. To investigate the tissue and cellular localization of HDA6 in roots, we generated Arabidopsis, rice, and cassava transformants expressing HDA6 fused to GFP under the control of its own promoter. We detected expression of HDA6-GFP in the nuclei of cells throughout the entire root in all three species (Fig. 2a, c, e). However, there was a difference among the species in the intracellular localization of HDA6-GFP: as well as expression in the nuclei, Arabidopsis and cassava also showed cytosolic localization of HDA6-GFP, while it was not obvious in rice (Fig. 2b, d, f). In addition, we found that in Arabidopsis only, the fusion protein clearly formed speckles in interphase nuclei of root meristem cells. AtHDA6-GFP speckles were not seen in elongating root cells (Fig. 2g, h), differentiated root cells (Fig. 2i), hypocotyl cells (Fig. 2j), or pavement cells in the leaf (Fig. 2k). These results suggested that AtHDA6 and MeHDA6 might have a cytosolic function, while AtHDA6 might have additional functions in the nuclei of root meristem cells besides the ones that OsHDA6-GFP and MeHDA6 have.
Expression patterns of HDA6 in Arabidopsis, rice, and cassava. a–k Observations of HDA6-GFP expression patterns in Arabidopsis (a, b, g–k), rice (c, d), and cassava (e, f). a–f root apical meristem cells, g, h elongating cell, i differentiated cell, j hypocotyl cell, k pavement cells of leaf. Scale bars: a, c, e, g, k = 50 μm; b, d, f, h–j = 10 μm
AtHDA6 actively localizes on chromocenters in Arabidopsis root meristem cells
It has been reported that the loss of function of athda6 causes defects in the organization and gene silencing of rDNA repeats (Earley et al. 2010; Probst et al. 2004). Considering this, we expected that AtHDA6-GFP would be accumulated around heterochromatic regions termed chromocenters, which contain centromeric and pericentromeric DNA arrays, and some of which also include rDNA repeats (Maluszynska and Heslopharrison 1991). To evaluate this possibility, nuclei of AtHDA6-GFP-expressing cells were counterstained with DAPI, enabling us to identify the chromocenters as DAPI-dense spots. Some of the AtHDA6-GFP speckles were indeed found to coincide with the positions of chromocenters (Fig. 3a). Next, we tested whether the colocalization of AtHDA6-GFP speckles and chromocenters is by chance, by comparing the theoretical colocalizing ratio that would be expected if AtHDA6-GFP speckle distribution were random with the ratio calculated from observed measurements (Fig. 3b). We found that the observed ratio of AtHDA6-GFP speckles colocalizing with chromocenters was significantly higher than the theoretical ratio, indicating that AtHDA6 actively localizes on chromocenters (Fig. 3c). To investigate in more detail the localization of AtHDA6 around chromocenters, we simultaneously visualized AtHDA6-GFP and rDNA or pericentromeric repeats by FISH. The results showed that some AtHDA6-GFP speckles coincided with both 5S rDNA, 45S rDNA and pericentromeric signals although the speckles preferentially colocalized with 45S rDNA (Fig. 3d). Taken together, these results suggested that AtHDA6 actively localizes on rDNA and pericentromeric loci and might regulate their organization in root meristem cells. To check the relationship between the formation of chromocenters and AtHDA6-GFP speckles, we observed chromocenters in Arabidopsis, rice, and cassava using DAPI staining (Fig. 3e). Chromocenters were seen in root meristem cells of all plants tested, although AtHDA6-GFP speckles were only seen in Arabidopsis, suggesting no causal relationship between the formation of chromocenters and AtHDA6-GFP speckles.
Analysis of cellular localization of AtHDA6 in Arabidopsis. a Observation of DAPI-stained chromocenter in root meristematic cells of AtHDA6-GFP-expressing plants. AtHDA6-GFP and DAPI staining are shown in green and magenta, respectively. The arrows indicate the AtHDA6 speckles colocalized with chromocenters. Scale bar = 5 μm. b Calculation of theoretical probability for random co-localization of chromocenter and AtHDA6-GFP. In order to calculate the probability that the regions of HDA6 speckle (dark green) and the chromocenters (dark magenta) were randomly colocalized, the nuclear area, radius of HDA6 speckles, and radius of chromocenters were measured from fluorescence images. If the HDA6 speckle was randomly distributed, the HDA6 speckle would be colocalized on the chromocenter with the probability: area of HDA6/(the nucleoplasm area excluding the nucleolus area). R = radius of HDA6 speckle + radius of chromocenter; then area of HDA6 (pale green area) = πR2. c Comparison between the theoretical ratio for a random distribution and the observed ratio of AtHDA6 speckles colocalizing with chromocenters. Mean ± SE, n = 40. *Significant difference, P < 0.01 (Student’s t test). d Observation of pericentromere (180 bp), 5S rDNA and 45S rDNA loci in AtHDA6-GFP-expressing plants. In upper panels, FISH, AtHDA6-GFP and DAPI for indicated loci are shown in red, green, and blue, respectively. The arrows indicate AtHDA6 speckles colocalized with pericentromere, 5S rDNA, or 45S rDNA loci. The arrowheads indicate the AtHDA6 speckles independent of pericentromere, 5S rDNA, or 45S rDNA loci. Scale bar = 5 μm. Quantification data of colocalization ratio between HDA6 speckles and FISH signals is shown in lower panel. Different alphabets indicate a significant difference, P < 0.05 (Tukey–Kramer). e Observation of DAPI-stained chromocenter in root meristematic cells of Arabidopsis, rice, and cassava. Scale bar = 10 μm (color figure online)
The extracellular pH changes within 20 min after acetic acid treatment
In response to drought stress, AtHDA6 acts as a switch to redirect the metabolic flux, leading to increased production of acetic acid in aerial parts that confers the plants with tolerance to drought stress. During this event, acetic acid is used as the substrate for histone hyperacetylation of genes, promoting jasmonic acid signaling that is required for tolerance of drought stress. Exogenous acetic acid treatment of plants further enhances their drought stress tolerance (Kim et al. 2017). Considering that the root is the organ affected first by drought stress and acetic acid treatment, it is important to understand the action of AtHDA6 in roots. However, the cellular dynamics of AtHDA6 responding to acetic acid treatment and also to drought stress are unknown.
To evaluate the dynamics of the response to acetic acid treatment, it is necessary to understand how long it takes for root cells to be exposed to the effects of acetic acid treatment. To determine such time-scales, we focused on pH changes, as it is known that the pH of xylem sap increases after treatment with acetic acid (Kim et al. 2017) or drought stress (Wilkinson and Davies 1997).
To assess pH changes in root tips, first we tried to establish a pH monitoring system using a microchannel device and the genetically-encoded pH biosensor pHusion (Gjetting et al. 2012). We created a microchannel device made of PDMS that was designed to cover a plant root on a glass slide (Fig. 4a). The dimensions of the microchannel are height 90 µm, width 3 mm, and length about 15 mm. In this microchannel device, the space for the plant root is filled with perfusing solution. The solution is easily removed from the outlet hole at the distal end of a plant root and a new solution injected via the inlet slit at the opposite end. The removal and injection of solution are performed simultaneously using a peristaltic pump. The width of the space for the plant root is narrow enough to limit the movement of the root. Therefore, we can perfuse the solution and observe the root using confocal laser microscopy without the need for any chemicals (such as poly-L-lysine or agar, which could inhibit plant growth) for stabilizing the root position during live imaging over some hours. Using this system, we succeeded in producing a 12-h time-course of images of the pH biosensor in a root without major changes in its positioning and during which time the root continued to grow (Fig. 4b).
Validation of the microchannel-device monitoring system. a Schematic of the microchannel device. b Twelve-hour time-lapse imaging of a single root expressing Apo-pHusion. Scale bar = 100 μm. c pH monitoring of a root exposed to a series of different pH buffers. The mature region of a root expressing Apo-pHusion was observed. The buffers were exchanged by perfusion using a peristaltic pump. The ratio of EGFP fluorescence intensity to mRFP1 fluorescence intensity in images is expressed using 16 colors. Scale bar = 100 μm. d pH monitoring in epidermal cells and vascular bundle cells in response to changes of external pH. The outermost layer and the fourth layer of cells were considered as epidermal cells and vascular bundle cells, respectively. The ratio of EGFP fluorescence intensity to mRFP1 fluorescence intensity at each pH is plotted. Mean ± SE, n = 3
Next, we checked whether pH monitoring is feasible in our imaging system. pHusion and Apo-pHusion is a ratio-metric pH biosensor system consisting of the tandem concatenation of the pH-sensitive enhanced green fluorescent protein (EGFP) and the pH-tolerant mRFP1 (Gjetting et al. 2012). By calculating the ratio of fluorescence intensity of EGFP against that of mRFP1, we can see the relative change in pH. When we observed roots of Apo-pHusion (which detects the pH change only in the apoplast; Gjetting et al. 2012) during perfusion with a series of pH buffers (pH 8–5), the ratio of fluorescence intensity gradually decreased around epidermal cells in response to the external pH changes (Fig. 4c, d). In contrast, the ratio did not change in the vascular bundle cells upon external pH change (Fig. 4c, d). The difference between epidermal cells and vascular bundle cells might be due to the existence of the casparian strip in the endodermal cell walls that prevents the flow of apoplastic solution. Taken together, we concluded that the pH biosensor could monitor the pH change under our imaging system.
Using our pH monitoring system, we analyzed the effect of acetic acid treatment on intra- and extra-cellular pH. To reproduce the conditions of acetic acid treatment that confers drought tolerance in soil culture (Kim et al. 2017), roots were treated with acetic acid soil extract and compared with treatment with a water-based soil extract. One-hour monitoring of Apo-pHusion revealed that apoplastic pH increased soon after the treatment with acetic acid soil extract, and then it reached a steady state from 20 to 60 min treatment. The water-based soil extract treatment did not cause such a rise in pH (Fig. 5a, c). In contrast, intracellular pH detected using pHusion was increased by either soil extract or acetic acid soil extract treatment and the extent of pH increase was similar between the treatments throughout the 1-h observation (Fig. 5b, d). In addition, a similar tendency was seen with the treatment of acetic acid buffers (Fig. S1). These results indicated that the effects of the acetic acid soil extract appeared within 20 min after the treatment, but only in the apoplastic fraction. Thus, it was suggested that acetic acid increases the pH of the root apoplast space.
pH monitoring in Arabidopsis root meristem after acetic acid treatment. a, b pH in Arabidopsis root meristem every 10 min after treatment with soil extract or acetic acid soil extract. The ratio-metric images of Apo-pHusion (a) and pHusion (b) are shown using 16 colors based on the ratio of fluorescence intensity of EGFP to mRFP1. White dotted lines indicate analyzed sites. Scale bar = 100 μm. The area surrounded by a white dotted line was analyzed in c, d. c, d The ratio of fluorescence intensity for Apo-pHusion (c) and pHusion (b) at each time point. Mean ± SE, n = 5
AtHDA6 localization changes in response to drought stress but not to acetic acid
Based on the results of pH monitoring, we evaluated the dynamics of AtHDA6 in interphase nuclei of root meristem cells 1 h after treatment with an acetic acid soil extract. In this analysis, we considered AtHDA6-GFP speckle formation as an indicator for evaluating the subnuclear accumulation of AtHDA6. We found that treatment with the acetic acid soil extract did not affect the frequency of AtHDA6-GFP speckle formation (Figs. 6a, S2a). In contrast, 1 h after mannitol treatment, which induces a drought response in plants, we found a significant increase of AtHDA6-GFP speckle formation (Fig. 6b). In addition, we found that even the treatment of mannitol did not cause clear HDA6-GFP speckle formation in cassava and rice (Fig. S3), further supporting the idea that AtHDA6 has different functions from HDA6 in cassava and rice. Furthermore, we examined the formation of AtHDA6 speckle after the treatment of an inhibitor of histone deacetylase, TSA (Fig. S2b). The treatment of TSA significantly reduced the number of AtHDA6-GFP speckles, suggesting that the speckle formation associates with the activity of histone deacetylase. This idea is supported by our observation, which even AtHDA6 speckle other than chromocenters exhibited lower H3K9ac level (Line 2 panel in Fig. S2c). Taken together, these results suggested that AtHDA6 activity is responsive to drought stress but not to those aspects of drought stress responses that are dependent on acetic acid in the root.
Effect of acetic acid and drought on AtHDA6 dynamics. a Effect of 1 h treatment with acetic acid soil extract on AtHDA6-GFP formation. Upper panel: representative images of a cell before and after acetic acid soil extract treatment. Lower panel: number of speckles per nucleus under the indicated conditions. Mean ± SE, n = 10. n.s., not significant (Student’s t test). b Effect of 1 h treatment with mannitol on AtHDA6-GFP formation. Upper panel: representative images of a cell before and after mannitol treatment. Lower panel: number of speckles per nucleus under the indicated conditions. Mean ± SE, n = 5. *Significant difference, P < 0.05 (Student’s t test)
Discussion
In this study, we established that a portion of AtHDA6 is localized on chromocenters, which contain a pericentromere, 5S and 45S rDNAs, transposable elements, and silenced genes. The fact that more than half of AtHDA6-GFP speckles were colocalized with chromocenters (Fig. 3) provokes a question: are AtHDA6 speckles colocalized with specific chromocenters? Given that 5S rDNAs are located on chromosomes 3, 4, and 5 and 45S rDNA are located on chromosomes 2 and 4 (Layat et al. 2012), AtHDA6 has a potential to predominantly form speckles at these chromosomes beside with chromosome 1 (Fig. 3d). In other words, it might be that an AtHDA6 speckle formation is not specific to a certain chromocenter. Further, what is the significance of AtHDA6 speckle formation? It has been suggested that AtHDA6 functions in the formation of chromocenters (Vaillant et al. 2007) and local heterochromatin organization in concert with DNA methyltransferase, MET1 (To et al. 2011). Consistently, AtHDA6 speckles were also seen in regions other than chromocenters (Fig. 3). The formation of AtHDA6 speckles might have roles in the organization of not only robust but also local and temporary heterochromatin. Interestingly, the AtHDA6-GFP speckles were only observed in meristematic cells (Fig. 2), implying some association between speckle formation and the characteristics of meristematic cells. Meristematic cells are known to show higher levels of histone acetylation compared with differentiated cells in Arabidopsis (Rosa et al. 2014). Indeed, we found that AtHDA6 were localized in the speckle and, consequently, the speckle exhibited lower H3K9ac level (Fig. S2b, c). Therefore, it is conceivable that AtHDA6-enriched speckle formation allows hypoacetylation of histones to organize local heterochromatin in an environment of higher concentration of acetylated histone. Unlike the case in Arabidopsis, HDA6-GFP was unlikely to form speckles in rice or cassava (Fig. 2), suggesting that AtHDA6 has a unique function among these plant species. This might be attributable to differences in genomic organization such as the genome-wide distribution and extent of histone acetylation.
It is reported that AtHDA6 associates with and negatively regulates drought-responsive genes under normal condition (Kim et al. 2017). When plants are exposed to drought stress, AtHDA6 becomes dissociated from the genes, allowing them to be acetylated and activated. Therefore, we predicted that the number of AtHDA6-GFP speckles would be decreased as genes became activated under drought stress conditions. Surprisingly, we found the number of speckles increased immediately after mannitol treatment to induce drought stress (Fig. 6b). This result implies that the activity of AtHDA6 is rapidly enhanced by drought stress to repress a certain set of genes that is also crucial for dealing with drought stress. In contrast to the results for mannitol treatment, the number of speckles was similar before and after acetic acid treatment (Figs. 6a, S2a), implying that the acetic acid treatment did not enhance AtHDA6 activity. Taken together, our results suggest that AtHDA6 plays a role in an upstream stage of the drought stress responses that are activated by increased production of acetic acid in root meristematic cells, similar to the case in aerial parts (Kim et al. 2017).
In order to investigate the effects of acetic acid treatment on intra- and extra-cellular pH, we developed a microchannel device that enabled us to observe living roots for more than 12 h under microscopy while changing the external solution. Adopting this perfusion system allowed us to investigate the response of certain proteins fused with fluorescent proteins to environmental stress in living roots. Similar system such as RootChip and TRIS has been developed previously (Grossmann et al. 2011; Massalha et al. 2017). However, those methods required large and expensive devices. Compared to those, our system is cheaper and smaller and has high versatility for the analysis of environmental responses evoked during liquid treatments of plant roots. If we assume that the fluorescence ratio in epidermal cells reflects the external pH, the pH in vascular bundle cells would be maintained at between pH 6 and 7 (Fig. 4c, d). But the calculated pH is inconsistent with the estimated apoplastic pH 5.3 under normal growth conditions (Felle 2001). Therefore, in order to measure the absolute pH, we should perform the calibration of pHusion in individual plants as shown in the previous report (Gjetting et al. 2012).
Using our system, we showed for the first time that treatment with acetic acid induced alkalinization of the apoplast in the root tip within 20 min (Figs. 5, S1). This finding is consistent with the observed increase in the pH of the xylem sap, whose content is part of the apoplast, after 1 h acetic acid treatment (Kim et al. 2017). Although the transport system for acetate in plants remains unknown, acetate ions (to which the plasma membrane is impermeable) are likely to be co-transported from the apoplast to the cytosol by a proton co-transporter, similar to the case for Cl− under salinity (Geilfus 2017). This would result in a decrease of the proton concentration in the apoplast and its consequent alkalinization (Geilfus 2017). At the same time, the cytosolic proton concentration would be expected to increase, resulting in a lower pH. However, contrary to our expectations, treatment with acetic acid soil extract also caused alkalinization in the cytosol (Figs. 5, S1). Considering that even the water-based soil extract or the buffer without acetic acid caused cytosolic alkalinization to a similar extent to the acetic acid treatment (Figs. 5, S1), we speculate that cytosolic alkalinization is mere a consequence of higher pH in the treated soil extract (pH 6.2 for water extract; pH 6.5 for acetic acid extract) or the buffers (pH 7.0) compared to that in pre-growing medium (pH 5.8). Under these conditions, the effect of increased proton concentration derived from acetate intake would be masked by cytosolic alkalinization. Alternatively, cytoplasmic chemicals, ions, and organelles (especially the vacuole) might rapidly buffer or sequestrate protons. In line with the result that acetic acid did not affect AtHDA6 accumulation in nuclei (Fig. 6), acetate in the roots might have a role in transducing the signal of drought stress from root to shoot through alkalinizing the apoplast soon after uptake by the plant. A certain amount of acetate not used for apoplastic alkalinization might be allocated to aerial parts, and there used as a substrate for histone acetylation (Kim et al. 2017).
References
Allis CD, Jenuwein T (2016) The molecular hallmarks of epigenetic control. Nat Rev Genet 17:487–500
Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21:381–395
Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS (2010) Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev 24:1119–1132
Felle HH (2001) pH: signal and messenger in plant cells. Plant Biol 3:577–591
Geilfus CM (2017) The pH of the apoplast: dynamic factor with functional impact under stress. Mol Plant 10:1371–1386
Gjetting SK, Ytting CK, Schulz A, Fuglsang AT (2012) Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J Exp Bot 63:3207–3218
Grossmann G, Guo WJ, Ehrhardt DW, Frommer WB, Sit RV, Quake SR, Meier M (2011) The RootChip: an integrated microfluidic chip for plant science. Plant Cell 23:4234–4240
Hasegawa J, Sakamoto Y, Nakagami S, Aida M, Sawa S, Matsunaga S (2016) Three-dimensional imaging of plant organs using a simple and rapid transparency technique. Plant Cell Physiol 57:462–472
Hasegawa J, Sakamoto T, Fujimoto S, Yamashita T, Suzuki T, Matsunaga S (2018) Auxin decreases chromatin accessibility through the TIRl/AFBs auxin signaling pathway in proliferative cells. Sci Rep 8:12
Hida H, Nishiyama H, Sawa S, Higashiyama T, Arata H (2015) Chemotaxis assay of plant-parasitic nematodes on a gel-filled microchannel device. Sens Actuat B-Chem 221:1483–1491
Hirakawa T, Hasegawa J, White CI, Matsunaga S (2017) RAD54 forms DNA repair foci in response to DNA damage in living plant cells. Plant J 90:372–382
Hollender C, Liu ZC (2008) Histone deacetylase genes in Arabidopsis development. J Integrat Plant Biol 50:875–885
Kim JM, To TK, Seki M (2012) An epigenetic integrator: new insights into genome regulation, environmental stress responses and developmental controls by histone deacetylase 6. Plant Cell Physiol 53:794–800
Kim JM, To TK, Matsui A, Tanoi K, Kobayashi NI, Matsuda F, Habu Y, Ogawa D, Sakamoto T, Matsunaga S, Bashir K, Rasheed S, Ando M, Takeda H, Kawaura K, Kusano M, Fukushima A, Endo TA, Kuromori T, Ishida J, Morosawa T, Tanaka M, Torii C, Takebayashi Y, Sakakibara H, Ogihara Y, Saito K, Shinozaki K, Devoto A, Seki M (2017) Acetate-mediated novel survival strategy against drought in plants. Nat Plants 3:7
Layat E, Sáez-Vásquez J, Tourmente S (2012) Regulation of Pol I-transcribed 45S rDNA and Pol III-transcribed 5S rDNA in Arabidopsis. Plant Cell Physiol 53:267–276
Maluszynska J, Heslopharrison JS (1991) Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. Plant J 1:159–166
Massalha H, Korenblum E, Malitsky S, Shapiro OH, Aharoni A (2017) Live imaging of root-bacteria interactions in a microfluidics setup. Proc Natl Acad Sci USA 14:4549–4554
Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, Watanabe Y, Nakamura K, Kimura T, Ishiguro S (2007) Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in Transgenic analysis of plants. Biosci Biotechnol Biochem 71:2095–2100
Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30:5036–5055
Park J, Kurihara D, Higashiyama T, Arata H (2014) Fabrication of microcage arrays to fix plant ovules for long-term live imaging and observation. Sens Actuat B-Chem 191:178–185
Pontes O, Lawrence RJ, Silva M, Preuss S, Costa-Nunes P, Earley K, Neves N, Viegas W, Pikaard CS (2007) Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS One 2:e1157
Probst AV, Fagard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, Vaucheret H, Scheid OM (2004) Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16:1021–1034
Rosa S, Ntoukakis V, Ohmido N, Pendle A, Abranches R, Shaw P (2014) Cell differentiation and development in Arabidopsis are associated with changes in histone dynamics at the single-cell level. Plant Cell 26:4821–4833
Sakamoto T, Inui YT, Sotta N, Hirakawa T, Matsunaga TM, Fukao Y, Matsunaga S, Fujiwara T (2018) Proteasomal degradation of BRAHMA promotes Boron tolerance in Arabidopsis. Nat Commun 9:5258
To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T, Kimura H, Yokoyama S, Shinozaki K, Seki M (2011) Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet 7:14
Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47:969–976
Utsumi Y, Utsumi C, Tanaka M, Ha VT, Matsui A, Takahashi S, Seki M (2017) Formation of friable embryogenic callus in cassava is enhanced under conditions of reduced nitrate, potassium and phosphate. PLoS One 12:e0180736
Vaillant I, Tutois S, Cuvillier C, Schubert I, Tourmente S (2007) Regulation of Arabidopsis thaliana 5S rRNA genes. Plant Cell Physiol 48:745–752
Wilkinson S, Davies WJ (1997) Xylem sap pH increase: a drought signal received at the apoplastic face of the guard cell that involves the suppression of saturable abscisic acid uptake by the epidermal symplast. Plant Physiol 113:559–573
Zentner GE, Henikoff S (2013) Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol 20:259–266
Acknowledgements
This research was supported by Japan Science and Technology Agency (JST) CREST Grant (JPMJCR13B4) and MXT/JSPS KAKENHI (15H05955, 15H05962 and 19H03259) to SM and by MXT/JSPS KAKENHI (18K14743) and the Sasakawa Scientific Research Grant from Japan Science Society to YS. We thank Huw Tyson, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kurita, K., Sakamoto, Y., Naruse, S. et al. Intracellular localization of histone deacetylase HDA6 in plants. J Plant Res 132, 629–640 (2019). https://doi.org/10.1007/s10265-019-01124-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-019-01124-8